ssDNA-dependent colocalization of adeno-associated virus Rep and herpes simplex virus ICP8 in nuclear replication domains
- PMID: 14576307
- PMCID: PMC275469
- DOI: 10.1093/nar/gkg827
ssDNA-dependent colocalization of adeno-associated virus Rep and herpes simplex virus ICP8 in nuclear replication domains
Abstract
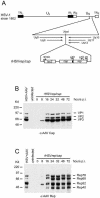
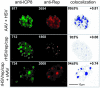
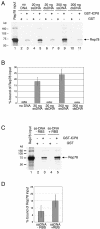
The subnuclear distribution of replication complex proteins is being recognized as an important factor for the control of DNA replication. Herpes simplex virus (HSV) single-strand (ss)DNA-binding protein, ICP8 (infected cell protein 8) accumulates in nuclear replication domains. ICP8 also serves as helper function for the replication of adeno-associated virus (AAV). Using quantitative 3D colocalization analysis we show that upon coinfection of AAV and HSV the AAV replication protein Rep and ICP8 co-reside in HSV replication domains. In contrast, Rep expressed by a recombinant HSV, in the absence of AAV DNA, displayed a nuclear distribution pattern distinct from that of ICP8. Colocal ization of Rep and ICP8 was restored by the reintroduction of single-stranded AAV vector genomes. In vitro, ICP8 displayed direct binding to Rep78. Single-stranded recombinant AAV DNA strongly stimulated this interaction, whereas double-stranded DNA was ineffective. Our findings suggest that ICP8 by its strong ssDNA-binding activity exploits the unique single-strandedness of the AAV genome to form a tripartite complex with Rep78 and AAV ssDNA. This novel mechanism for recruiting components of a functional replication complex directs AAV to subnuclear HSV replication compartments where the HSV replication complex can replicate the AAV genome.
Figures
Similar articles
-
DNA-binding activity of adeno-associated virus Rep is required for inverted terminal repeat-dependent complex formation with herpes simplex virus ICP8.J Virol. 2012 Mar;86(5):2859-63. doi: 10.1128/JVI.06364-11. Epub 2011 Dec 28. J Virol. 2012. PMID: 22205745 Free PMC article.
-
The Rep protein of adeno-associated virus type 2 interacts with single-stranded DNA-binding proteins that enhance viral replication.J Virol. 2004 Jan;78(1):441-53. doi: 10.1128/jvi.78.1.441-453.2004. J Virol. 2004. PMID: 14671124 Free PMC article.
-
Role of the herpes simplex virus helicase-primase complex during adeno-associated virus DNA replication.J Virol. 2006 Jun;80(11):5241-50. doi: 10.1128/JVI.02718-05. J Virol. 2006. PMID: 16699004 Free PMC article.
-
Definition of herpes simplex virus helper functions for the replication of adeno-associated virus type 5.J Gen Virol. 2015 Apr;96(Pt 4):840-850. doi: 10.1099/vir.0.000034. Epub 2014 Dec 22. J Gen Virol. 2015. PMID: 25535322
-
Chimeric herpes simplex virus/adeno-associated virus amplicon vectors.Curr Gene Ther. 2006 Jun;6(3):315-24. doi: 10.2174/156652306777592090. Curr Gene Ther. 2006. PMID: 16787183 Review.
Cited by
-
Roles of E4orf6 and VA I RNA in adenovirus-mediated stimulation of human parvovirus B19 DNA replication and structural gene expression.J Virol. 2012 May;86(9):5099-109. doi: 10.1128/JVI.06991-11. Epub 2012 Feb 22. J Virol. 2012. PMID: 22357277 Free PMC article.
-
Fluorescence Microscopy in Adeno-Associated Virus Research.Viruses. 2023 May 16;15(5):1174. doi: 10.3390/v15051174. Viruses. 2023. PMID: 37243260 Free PMC article. Review.
-
Herpes simplex virus type 1/adeno-associated virus hybrid vectors.Open Virol J. 2010 Jun 18;4:109-22. doi: 10.2174/1874357901004030109. Open Virol J. 2010. PMID: 20811580 Free PMC article.
-
Identification of rep-associated factors in herpes simplex virus type 1-induced adeno-associated virus type 2 replication compartments.J Virol. 2010 Sep;84(17):8871-87. doi: 10.1128/JVI.00725-10. Epub 2010 Jun 23. J Virol. 2010. PMID: 20573815 Free PMC article.
-
Human Bocavirus 1 NP1 acts as an ssDNA-binding protein to help AAV2 DNA replication and cooperates with RPA to regulate AAV2 capsid expression.J Virol. 2024 Mar 19;98(3):e0151523. doi: 10.1128/jvi.01515-23. Epub 2024 Feb 7. J Virol. 2024. PMID: 38323812 Free PMC article.
References
-
- Conway J., Rhys,C., Zolotukhin,I., Zolotukhin,S., Muzyczka,N., Hayward,G. and Byrne,B. (1999) High-titer recombinant adeno-associated virus production utilizing a recombinant herpes simplex virus type I vector expressing AAV-2 rep and cap. Gene Ther., 6, 986–993. - PubMed
-
- Muzyczka N. and Berns,K.I. (2001) Parvoviridae: the viruses and their replication. In Knipe,D.M. and Howley,P.M. (eds), Fields Virology. Lippincott, Philadelphia, PA, Vol. 2, pp. 2327–2359.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

