rsly1 binding to syntaxin 5 is required for endoplasmic reticulum-to-Golgi transport but does not promote SNARE motif accessibility
- PMID: 14565970
- PMCID: PMC307537
- DOI: 10.1091/mbc.e03-07-0535
rsly1 binding to syntaxin 5 is required for endoplasmic reticulum-to-Golgi transport but does not promote SNARE motif accessibility
Abstract
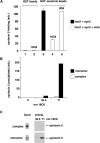
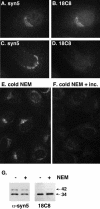
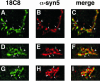
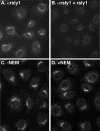
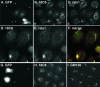
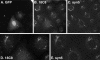
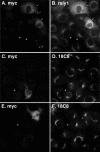
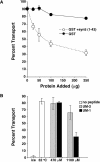
Although some of the principles of N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) function are well understood, remarkably little detail is known about sec1/munc18 (SM) protein function and its relationship to SNAREs. Popular models of SM protein function hold that these proteins promote or maintain an open and/or monomeric pool of syntaxin molecules available for SNARE complex formation. To address the functional relationship of the mammalian endoplasmic reticulum/Golgi SM protein rsly1 and its SNARE binding partner syntaxin 5, we produced a conformation-specific monoclonal antibody that binds only the available, but not the cis-SNARE-complexed nor intramolecularly closed form of syntaxin 5. Immunostaining experiments demonstrated that syntaxin 5 SNARE motif availability is nonuniformly distributed and focally regulated. In vitro endoplasmic reticulum-to-Golgi transport assays revealed that rsly1 was acutely required for transport, and that binding to syntaxin 5 was absolutely required for its function. Finally, manipulation of rsly1-syntaxin 5 interactions in vivo revealed that they had remarkably little impact on the pool of available syntaxin 5 SNARE motif. Our results argue that although rsly1 does not seem to regulate the availability of syntaxin 5, its function is intimately associated with syntaxin binding, perhaps promoting a later step in SNARE complex formation or function.
Figures




Similar articles
-
Mammalian Sly1 regulates syntaxin 5 function in endoplasmic reticulum to Golgi transport.J Biol Chem. 1996 Jul 5;271(27):15866-9. doi: 10.1074/jbc.271.27.15866. J Biol Chem. 1996. PMID: 8663406
-
Three-dimensional structure of the rSly1 N-terminal domain reveals a conformational change induced by binding to syntaxin 5.J Mol Biol. 2005 Feb 18;346(2):589-601. doi: 10.1016/j.jmb.2004.12.004. Epub 2004 Dec 24. J Mol Biol. 2005. PMID: 15670607
-
Sly1 protein bound to Golgi syntaxin Sed5p allows assembly and contributes to specificity of SNARE fusion complexes.J Cell Biol. 2002 May 13;157(4):645-55. doi: 10.1083/jcb.200202006. Epub 2002 May 6. J Cell Biol. 2002. PMID: 11994317 Free PMC article.
-
Stx5-Mediated ER-Golgi Transport in Mammals and Yeast.Cells. 2019 Jul 26;8(8):780. doi: 10.3390/cells8080780. Cells. 2019. PMID: 31357511 Free PMC article. Review.
-
SNAREs and membrane fusion in the Golgi apparatus.Biochim Biophys Acta. 1998 Aug 14;1404(1-2):9-31. doi: 10.1016/s0167-4889(98)00044-5. Biochim Biophys Acta. 1998. PMID: 9714710 Review.
Cited by
-
Sec24C/D-isoform-specific sorting of the preassembled ER-Golgi Q-SNARE complex.Mol Biol Cell. 2016 Sep 1;27(17):2697-707. doi: 10.1091/mbc.E16-04-0229. Epub 2016 Jul 13. Mol Biol Cell. 2016. PMID: 27413010 Free PMC article.
-
Munc18-1 mutations that strongly impair SNARE-complex binding support normal synaptic transmission.EMBO J. 2012 May 2;31(9):2156-68. doi: 10.1038/emboj.2012.72. Epub 2012 Mar 23. EMBO J. 2012. PMID: 22446389 Free PMC article.
-
An imaging-based RNA interference screen for modulators of the Rab6-mediated Golgi-to-ER pathway in mammalian cells.Front Cell Dev Biol. 2022 Nov 29;10:1050190. doi: 10.3389/fcell.2022.1050190. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36523508 Free PMC article.
-
The SM protein Sly1 accelerates assembly of the ER-Golgi SNARE complex.Proc Natl Acad Sci U S A. 2014 Sep 23;111(38):13828-33. doi: 10.1073/pnas.1408254111. Epub 2014 Sep 4. Proc Natl Acad Sci U S A. 2014. PMID: 25189771 Free PMC article.
-
UNC-18 promotes both the anterograde trafficking and synaptic function of syntaxin.Mol Biol Cell. 2008 Sep;19(9):3836-46. doi: 10.1091/mbc.e08-02-0160. Epub 2008 Jul 2. Mol Biol Cell. 2008. PMID: 18596236 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

