Mitochondria use different mechanisms for transport of multispanning membrane proteins through the intermembrane space
- PMID: 14560025
- PMCID: PMC207575
- DOI: 10.1128/MCB.23.21.7818-7828.2003
Mitochondria use different mechanisms for transport of multispanning membrane proteins through the intermembrane space
Abstract
The mitochondrial inner membrane contains numerous multispanning integral proteins. The precursors of these hydrophobic proteins are synthesized in the cytosol and therefore have to cross the mitochondrial outer membrane and intermembrane space to reach the inner membrane. While the import pathways of noncleavable multispanning proteins, such as the metabolite carriers, have been characterized in detail by the generation of translocation intermediates, little is known about the mechanism by which cleavable preproteins of multispanning proteins, such as Oxa1, are transferred from the outer membrane to the inner membrane. We have identified a translocation intermediate of the Oxa1 preprotein in the translocase of the outer membrane (TOM) and found that there are differences from the import mechanisms of carrier proteins. The intermembrane space domain of the receptor Tom22 supports the stabilization of the Oxa1 intermediate. Transfer of the Oxa1 preprotein to the inner membrane is not affected by inactivation of the soluble TIM complexes. Both the inner membrane potential and matrix heat shock protein 70 are essential to release the preprotein from the TOM complex, suggesting a close functional cooperation of the TOM complex and the presequence translocase of the inner membrane. We conclude that mitochondria employ different mechanisms for translocation of multispanning proteins across the aqueous intermembrane space.
Figures
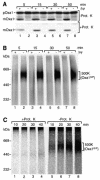
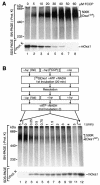
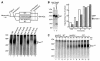

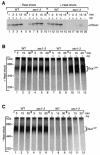

Similar articles
-
The mitochondrial import machinery for preproteins.Crit Rev Biochem Mol Biol. 2001;36(3):291-336. doi: 10.1080/20014091074200. Crit Rev Biochem Mol Biol. 2001. PMID: 11450972 Review.
-
Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein.J Biol Chem. 1999 Jun 4;274(23):16522-30. doi: 10.1074/jbc.274.23.16522. J Biol Chem. 1999. PMID: 10347216
-
Mitochondrial translocation contact sites: separation of dynamic and stabilizing elements in formation of a TOM-TIM-preprotein supercomplex.EMBO J. 2003 Oct 15;22(20):5370-81. doi: 10.1093/emboj/cdg532. EMBO J. 2003. PMID: 14532110 Free PMC article.
-
The import route of ADP/ATP carrier into mitochondria separates from the general import pathway of cleavable preproteins at the trans side of the outer membrane.J Biol Chem. 1998 Jun 26;273(26):16374-81. doi: 10.1074/jbc.273.26.16374. J Biol Chem. 1998. PMID: 9632701
-
Mechanisms of protein translocation into mitochondria.Biochim Biophys Acta. 1999 Nov 16;1422(3):235-54. doi: 10.1016/s0304-4157(99)00007-6. Biochim Biophys Acta. 1999. PMID: 10548718 Review.
Cited by
-
Mitochondrial Dynamics during Development.Newborn (Clarksville). 2023 Jan-Mar;2(1):19-44. doi: 10.5005/jp-journals-11002-0053. Epub 2023 Apr 6. Newborn (Clarksville). 2023. PMID: 37206581 Free PMC article.
-
The type III system-secreted effector EspZ localizes to host mitochondria and interacts with the translocase of inner mitochondrial membrane 17b.Infect Immun. 2011 Dec;79(12):4784-90. doi: 10.1128/IAI.05761-11. Epub 2011 Sep 26. Infect Immun. 2011. PMID: 21947777 Free PMC article.
-
Presequence recognition by the tom40 channel contributes to precursor translocation into the mitochondrial matrix.Mol Cell Biol. 2014 Sep 15;34(18):3473-85. doi: 10.1128/MCB.00433-14. Epub 2014 Jul 7. Mol Cell Biol. 2014. PMID: 25002531 Free PMC article.
-
Mitochondrial protein synthesis, import, and assembly.Genetics. 2012 Dec;192(4):1203-34. doi: 10.1534/genetics.112.141267. Genetics. 2012. PMID: 23212899 Free PMC article.
-
New component of ESCRT-I regulates endosomal sorting complex assembly.J Cell Biol. 2006 Dec 4;175(5):815-23. doi: 10.1083/jcb.200608053. J Cell Biol. 2006. PMID: 17145965 Free PMC article.
References
-
- Bauer, M., M. Behrens, K. Esser, G. Michaelis, and E. Pratje. 1994. PET1402, a nuclear gene required for proteolytic processing of cytochrome oxidase subunit 2 in yeast. Mol. Gen. Genet. 245:272-278. - PubMed
-
- Bauer, M. F., S. Hofmann, W. Neupert, and M. Brunner. 2000. Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol. 10:25-31. - PubMed
-
- Bauer, M. F., C. Sirrenberg, W. Neupert, and M. Brunner. 1996. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell 87:33-41. - PubMed
-
- Beddoe, T., and T. Lithgow. 2002. Delivery of nascent polypeptides to the mitochondrial surface. Biochim. Biophys. Acta 1592:35-39. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
