Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes
- PMID: 14555699
- PMCID: PMC280575
- DOI: 10.1105/tpc.016238
Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes
Erratum in
- Plant Cell. 2004 Feb;16(2):555
Abstract
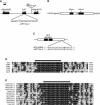
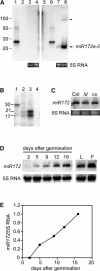
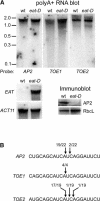
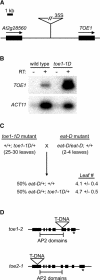
MicroRNAs (miRNAs) are approximately 21-nucleotide noncoding RNAs that have been identified in both animals and plants. Although in animals there is direct evidence implicating particular miRNAs in the control of developmental timing, to date it is not known whether plant miRNAs also play a role in regulating temporal transitions. Through an activation-tagging approach, we demonstrate that miRNA 172 (miR172) causes early flowering and disrupts the specification of floral organ identity when overexpressed in Arabidopsis. miR172 normally is expressed in a temporal manner, consistent with its proposed role in flowering time control. The regulatory target of miR172 is a subfamily of APETALA2 (AP2) transcription factor genes. We present evidence that miR172 downregulates these target genes by a translational mechanism rather than by RNA cleavage. Gain-of-function and loss-of-function analyses indicate that two of the AP2-like target genes normally act as floral repressors, supporting the notion that miR172 regulates flowering time by downregulating AP2-like target genes.
Figures

Similar articles
-
Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana.Plant Mol Biol. 2006 Jul;61(4-5):781-93. doi: 10.1007/s11103-006-0049-0. Plant Mol Biol. 2006. PMID: 16897492 Free PMC article.
-
The miR172 target TOE3 represses AGAMOUS expression during Arabidopsis floral patterning.Plant Sci. 2014 Feb;215-216:29-38. doi: 10.1016/j.plantsci.2013.10.010. Epub 2013 Oct 25. Plant Sci. 2014. PMID: 24388512
-
Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis.Plant J. 2012 May;70(4):549-61. doi: 10.1111/j.1365-313X.2012.04919.x. Epub 2012 Mar 5. Plant J. 2012. PMID: 22268548
-
Regulation of flowering time and floral patterning by miR172.J Exp Bot. 2011 Jan;62(2):487-95. doi: 10.1093/jxb/erq295. Epub 2010 Oct 15. J Exp Bot. 2011. PMID: 20952628 Review.
-
Regulated RNA processing in the control of Arabidopsis flowering.Int J Dev Biol. 2005;49(5-6):773-80. doi: 10.1387/ijdb.051995vq. Int J Dev Biol. 2005. PMID: 16096981 Review.
Cited by
-
miR172b controls the transition to autotrophic development inhibited by ABA in Arabidopsis.PLoS One. 2013 May 23;8(5):e64770. doi: 10.1371/journal.pone.0064770. Print 2013. PLoS One. 2013. PMID: 23717657 Free PMC article.
-
Structural determinants of miR156a precursor processing in temperature-responsive flowering in Arabidopsis.J Exp Bot. 2016 Aug;67(15):4659-70. doi: 10.1093/jxb/erw248. Epub 2016 Jun 21. J Exp Bot. 2016. PMID: 27335452 Free PMC article.
-
The micro-RNA72c-APETALA2-1 node as a key regulator of the common bean-Rhizobium etli nitrogen fixation symbiosis.Plant Physiol. 2015 May;168(1):273-91. doi: 10.1104/pp.114.255547. Epub 2015 Mar 4. Plant Physiol. 2015. PMID: 25739700 Free PMC article.
-
Rice osa-miR171c Mediates Phase Change from Vegetative to Reproductive Development and Shoot Apical Meristem Maintenance by Repressing Four OsHAM Transcription Factors.PLoS One. 2015 May 29;10(5):e0125833. doi: 10.1371/journal.pone.0125833. eCollection 2015. PLoS One. 2015. PMID: 26023934 Free PMC article.
-
Upregulation of LINC-AP2 is negatively correlated with AP2 gene expression with Turnip crinkle virus infection in Arabidopsis thaliana.Plant Cell Rep. 2016 Nov;35(11):2257-2267. doi: 10.1007/s00299-016-2032-9. Epub 2016 Jul 29. Plant Cell Rep. 2016. PMID: 27473526
References
-
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1995). Current Protocols in Molecular Biology. (New York: John Wiley & Sons).
-
- Blazquez, M.A., Ahn, J.H., and Weigel, D. (2003). A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet. 33, 168–171. - PubMed
-
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20. - PubMed
-
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. (2003). bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

