Novel simian homologues of Epstein-Barr virus
- PMID: 12970457
- PMCID: PMC228477
- DOI: 10.1128/jvi.77.19.10695-10699.2003
Novel simian homologues of Epstein-Barr virus
Abstract
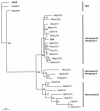
Thirty different lymphocryptoviruses (LCV), 26 of them novel, were detected in primates by a panherpesvirus PCR assay. Nineteen LCV from chimpanzees, bonobos, gorillas, and other Old World primates were closely related to Epstein-Barr virus (EBV), the type species of the genus lymphocryptovirus. Seven LCV originating from New World primates were related to callitrichine herpesvirus 3 (CalHV-3), the first recognized New World LCV. Importantly, a second LCV from gorillas and three LCV from orangutans and gibbons were only distantly related to EBV and CalHV-3. They were tentatively assigned to a novel genogroup of Old World primate LCV. The work described in the paper may also help identify an as yet unknown human LCV.
Figures

Similar articles
-
Complete genomic sequence of an Epstein-Barr virus-related herpesvirus naturally infecting a new world primate: a defining point in the evolution of oncogenic lymphocryptoviruses.J Virol. 2002 Dec;76(23):12055-68. doi: 10.1128/jvi.76.23.12055-12068.2002. J Virol. 2002. PMID: 12414947 Free PMC article.
-
Evolution of two types of rhesus lymphocryptovirus similar to type 1 and type 2 Epstein-Barr virus.J Virol. 1999 Nov;73(11):9206-12. doi: 10.1128/JVI.73.11.9206-9212.1999. J Virol. 1999. PMID: 10516028 Free PMC article.
-
Lymphocryptovirus phylogeny and the origins of Epstein-Barr virus.J Gen Virol. 2010 Mar;91(Pt 3):630-42. doi: 10.1099/vir.0.017251-0. Epub 2009 Nov 18. J Gen Virol. 2010. PMID: 19923263
-
Simian homologues of Epstein-Barr virus.Philos Trans R Soc Lond B Biol Sci. 2001 Apr 29;356(1408):489-97. doi: 10.1098/rstb.2000.0776. Philos Trans R Soc Lond B Biol Sci. 2001. PMID: 11313007 Free PMC article. Review.
-
Non-human Primate Lymphocryptoviruses: Past, Present, and Future.Curr Top Microbiol Immunol. 2015;391:385-405. doi: 10.1007/978-3-319-22834-1_13. Curr Top Microbiol Immunol. 2015. PMID: 26428382 Review.
Cited by
-
Endemic Viruses of Squirrel Monkeys (Saimiri spp.).Comp Med. 2015 Jun;65(3):232-40. Comp Med. 2015. PMID: 26141448 Free PMC article. Review.
-
African great apes are naturally infected with roseoloviruses closely related to human herpesvirus 7.J Virol. 2014 Nov;88(22):13212-20. doi: 10.1128/JVI.01490-14. Epub 2014 Sep 3. J Virol. 2014. PMID: 25187544 Free PMC article.
-
Novel mammalian herpesviruses and lineages within the Gammaherpesvirinae: cospeciation and interspecies transfer.J Virol. 2008 Apr;82(7):3509-16. doi: 10.1128/JVI.02646-07. Epub 2008 Jan 23. J Virol. 2008. PMID: 18216123 Free PMC article.
-
Identification and functional comparison of seven-transmembrane G-protein-coupled BILF1 receptors in recently discovered nonhuman primate lymphocryptoviruses.J Virol. 2015 Feb;89(4):2253-67. doi: 10.1128/JVI.02716-14. Epub 2014 Dec 10. J Virol. 2015. PMID: 25505061 Free PMC article.
-
Condylomatous genital lesions in cynomolgus macaques from Mauritius.Toxicol Pathol. 2013 Aug;41(6):893-901. doi: 10.1177/0192623312467521. Epub 2012 Dec 21. Toxicol Pathol. 2013. PMID: 23262641 Free PMC article.
References
-
- Ablashi, D. V., J. M. Easton, and J. H. Guegan. 1976. Herpesviruses and cancer in man and subhuman primates. Biomedicine 24:286-305. - PubMed
-
- Barahona, H., L. V. Melendez, and J. L. Melnick. 1974. A compendium of herpesviruses from non-human primates. Intervirology 3:175-192. - PubMed
-
- Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. - PubMed
-
- Chmielewicz, B., M. Goltz, and B. Ehlers. 2001. Detection and multigenic characterization of a novel gammaherpesvirus in goats. Virus Res. 75:87-94. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

