DNA topoisomerase III from the hyperthermophilic archaeon Sulfolobus solfataricus with specific DNA cleavage activity
- PMID: 12949102
- PMCID: PMC193750
- DOI: 10.1128/JB.185.18.5500-5507.2003
DNA topoisomerase III from the hyperthermophilic archaeon Sulfolobus solfataricus with specific DNA cleavage activity
Abstract
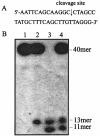
We report the production, purification, and characterization of a type IA DNA topoisomerase, previously designated topoisomerase I, from the hyperthermophilic archaeon Sulfolobus solfataricus. The protein was capable of relaxing negatively supercoiled DNA at 75 degrees C in the presence of Mg2+. Mutation of the putative active site Tyr318 to Phe318 led to the inactivation of the protein. The S. solfataricus enzyme cleaved oligonucleotides in a sequence-specific fashion. The cleavage occurred only in the presence of a divalent cation, preferably Mg2+. The cofactor requirement of the enzyme was partially satisfied by Cu2+, Co2+, Mn2+, Ca2+, or Ni2+. It appears that the enzyme is active with a broader spectrum of metal cofactors in DNA cleavage than in DNA relaxation (Mg2+ and Ca2+). The enzyme-catalyzed oligonucleotide cleavage required at least 7 bases upstream and 2 bases downstream of the cleavage site. Analysis of cleavage by the S. solfataricus enzyme on a set of oligonucleotides revealed a consensus cleavage sequence of the enzyme: 5'-G(A/T)CA(T)AG(T)G(A)X / XX-3'. This sequence bears more resemblance to the preferred cleavage sites of topoisomerases III than to those of topoisomerases I. Based on these data and sequence analysis, we designate the enzyme S. solfataricus topoisomerase III.
Figures

Similar articles
-
TopA, the Sulfolobus solfataricus topoisomerase III, is a decatenase.Nucleic Acids Res. 2018 Jan 25;46(2):861-872. doi: 10.1093/nar/gkx1247. Nucleic Acids Res. 2018. PMID: 29253195 Free PMC article.
-
Oligonucleotide cleavage and rejoining by topoisomerase III from the hyperthermophilic archaeon Sulfolobus solfataricus: temperature dependence and strand annealing-promoted DNA religation.Mol Microbiol. 2006 May;60(3):783-94. doi: 10.1111/j.1365-2958.2006.05133.x. Mol Microbiol. 2006. PMID: 16629677
-
Kinetic insights into the temperature dependence of DNA strand cleavage and religation by topoisomerase III from the hyperthermophile Sulfolobus solfataricus.Sci Rep. 2017 Jul 14;7(1):5494. doi: 10.1038/s41598-017-05837-5. Sci Rep. 2017. PMID: 28710489 Free PMC article.
-
A carboxylesterase from the hyperthermophilic archaeon Sulfolobus solfataricus: cloning of the gene, characterization of the protein.Gene. 2002 Jan 23;283(1-2):107-15. doi: 10.1016/s0378-1119(01)00879-4. Gene. 2002. PMID: 11867217
-
Bacterial topoisomerase I as a target for discovery of antibacterial compounds.Nucleic Acids Res. 2009 Feb;37(3):731-7. doi: 10.1093/nar/gkn936. Epub 2008 Nov 28. Nucleic Acids Res. 2009. PMID: 19042977 Free PMC article. Review.
Cited by
-
Archaea: A Gold Mine for Topoisomerase Diversity.Front Microbiol. 2021 May 25;12:661411. doi: 10.3389/fmicb.2021.661411. eCollection 2021. Front Microbiol. 2021. PMID: 34113328 Free PMC article. Review.
-
Identification and characterization of a highly conserved crenarchaeal protein lysine methyltransferase with broad substrate specificity.J Bacteriol. 2012 Dec;194(24):6917-26. doi: 10.1128/JB.01535-12. Epub 2012 Oct 19. J Bacteriol. 2012. PMID: 23086207 Free PMC article.
-
Calcium-driven DNA synthesis by a high-fidelity DNA polymerase.Nucleic Acids Res. 2017 Dec 1;45(21):12425-12440. doi: 10.1093/nar/gkx927. Nucleic Acids Res. 2017. PMID: 29040737 Free PMC article.
-
TopA, the Sulfolobus solfataricus topoisomerase III, is a decatenase.Nucleic Acids Res. 2018 Jan 25;46(2):861-872. doi: 10.1093/nar/gkx1247. Nucleic Acids Res. 2018. PMID: 29253195 Free PMC article.
-
Structural and biochemical basis for DNA and RNA catalysis by human Topoisomerase 3β.Nat Commun. 2022 Aug 9;13(1):4656. doi: 10.1038/s41467-022-32221-3. Nat Commun. 2022. PMID: 35945419 Free PMC article.
References
-
- Bergerat, A., B. de Massy, D. Gadelle, P. C. Varoutas, A. Nicolas, and P. Forterre. 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386:414-417. - PubMed
-
- Bhaduri, T., T. K. Bagui, D. Sikder, and V. Nagaraja. 1998. DNA topoisomerase I from Mycobacterium smegmatis. An enzyme with distinct features. J. Biol. Chem. 273:13925-13932. - PubMed
-
- Champoux, J. J. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70:369-413. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

