Human T-cell leukemia virus type 1 envelope glycoprotein gp46 interacts with cell surface heparan sulfate proteoglycans
- PMID: 12941902
- PMCID: PMC224595
- DOI: 10.1128/jvi.77.18.9922-9930.2003
Human T-cell leukemia virus type 1 envelope glycoprotein gp46 interacts with cell surface heparan sulfate proteoglycans
Abstract
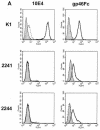
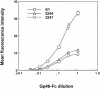
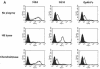
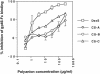
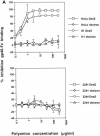
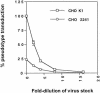
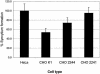
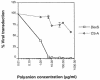
The major receptors required for attachment and entry of the human T-cell leukemia virus type 1 (HTLV-1) remain to be identified. Here we demonstrate that a functional, soluble form of the HTLV-1 surface envelope glycoprotein, gp46, fused to an immunoglobulin Fc region (gp46-Fc) binds to heparan sulfate proteoglycans (HSPGs) on mammalian cells. Substantial binding of gp46-Fc to HeLa and Chinese hamster ovary (CHO) K1 cells that express HSPGs was detected, whereas binding to the sister CHO lines 2244, which expresses no HSPGs, and 2241, which expresses no glycosaminoglycans (GAGs), was much reduced. Enzymatic removal of HSPGs from HeLa and CHO K1 cells also reduced gp46-Fc binding. Dextran sulfate inhibited gp46-Fc binding to HSPG-expressing cells in a dose-dependent manner, whereas chondroitin sulfate was less effective. By contrast, dextran sulfate inhibited gp46-Fc binding to GAG-negative cells such as CHO 2244, CHO 2241, and Jurkat T cells weakly or not at all. Dextran sulfate inhibited HTLV-1 envelope glycoprotein (Env)-pseudotyped virus infection of permissive, HSPG-expressing target cells and blocked syncytium formation between HTLV-1 Env-expressing cells and HSPG-expressing permissive target cells. Finally, HSPG-expressing cells were more permissive for HTLV-1 Env-pseudotyped virus infection than HSPG-negative cells. Thus, similar to other pathogenic viruses, HTLV-1 may have evolved to use HSPGs as cellular attachment receptors to facilitate its propagation.
Figures



Similar articles
-
Molecular aspects of HTLV-1 entry: functional domains of the HTLV-1 surface subunit (SU) and their relationships to the entry receptors.Viruses. 2011 Jun;3(6):794-810. doi: 10.3390/v3060794. Epub 2011 Jun 15. Viruses. 2011. PMID: 21994754 Free PMC article. Review.
-
Human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2 use different receptor complexes to enter T cells.J Virol. 2006 Sep;80(17):8291-302. doi: 10.1128/JVI.00389-06. J Virol. 2006. PMID: 16912281 Free PMC article.
-
Antibodies to the envelope glycoprotein of human T cell leukemia virus type 1 robustly activate cell-mediated cytotoxic responses and directly neutralize viral infectivity at multiple steps of the entry process.J Immunol. 2011 Jul 1;187(1):361-71. doi: 10.4049/jimmunol.1100070. Epub 2011 Jun 6. J Immunol. 2011. PMID: 21646298
-
Heparan sulfate proteoglycans mediate attachment and entry of human T-cell leukemia virus type 1 virions into CD4+ T cells.J Virol. 2005 Oct;79(20):12692-702. doi: 10.1128/JVI.79.20.12692-12702.2005. J Virol. 2005. PMID: 16188972 Free PMC article.
-
Molecular mechanisms affecting HTLV type 1-dependent fusion at the cell membrane: implications for inhibiting viral transmission.AIDS Res Hum Retroviruses. 2000 Nov 1;16(16):1731-6. doi: 10.1089/08892220050193227. AIDS Res Hum Retroviruses. 2000. PMID: 11080818 Review.
Cited by
-
Dynamic acquisition of HTLV-1 tax protein by mononuclear phagocytes: Role in neurologic disease.J Neuroimmunol. 2017 Mar 15;304:43-50. doi: 10.1016/j.jneuroim.2016.09.014. Epub 2016 Oct 3. J Neuroimmunol. 2017. PMID: 27769524 Free PMC article.
-
Heparan sulfate is an attachment factor for foamy virus entry.J Virol. 2012 Sep;86(18):10028-35. doi: 10.1128/JVI.00051-12. Epub 2012 Jul 11. J Virol. 2012. PMID: 22787203 Free PMC article.
-
Alternate receptor usage of neuropilin-1 and glucose transporter protein 1 by the human T cell leukemia virus type 1.Virology. 2010 Jan 20;396(2):203-12. doi: 10.1016/j.virol.2009.10.015. Epub 2009 Nov 13. Virology. 2010. PMID: 19913864 Free PMC article.
-
The Past, Present, and Future of a Human T-Cell Leukemia Virus Type 1 Vaccine.Front Microbiol. 2022 May 4;13:897346. doi: 10.3389/fmicb.2022.897346. eCollection 2022. Front Microbiol. 2022. PMID: 35602078 Free PMC article. Review.
-
Molecular aspects of HTLV-1 entry: functional domains of the HTLV-1 surface subunit (SU) and their relationships to the entry receptors.Viruses. 2011 Jun;3(6):794-810. doi: 10.3390/v3060794. Epub 2011 Jun 15. Viruses. 2011. PMID: 21994754 Free PMC article. Review.
References
-
- Bernfield, M., M. Gotte, P. Park, O. Reizes, M. Fitzgerald, J. Lincecum, and M. Zako. 1999. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 68:729-777. - PubMed
-
- Bobardt, M., A. Saphire, H.-C. Hung, X. Yu, B. Van der Schueren, Z. Zhang, G. David, and P. Gallay. 2003. Syndecan captures, protects and transmits HIV to T lymphocytes. Immunity 18:27-39. - PubMed
-
- Clapham, P., K. Nagy, P. Cheingsong, M. Exley, and R. Weiss. 1983. Productive infection and cell-free transmission of human T cell leukemia virus in a njonlymphoid cell line. Science 222:1125-1127. - PubMed
-
- Clasper, S., S. Vekemans, M. Fiore, M. Plebanski, P. Wordsworth, G. David, and D. Jackson. 1999. Inducible expression of the cell surface heparan sulfate proteoglycan syndecan-2 (fibroglycan) on human activated macrophages can regulate fibroblast growth factor action. J. Biol. Chem. 274:24113-24123. - PubMed
-
- Daenke, S., S. McCracken, and S. Booth. 1999. Hum. T cell leukemia/lymphoma virus type 1 syncytium formation is regulated in a cell specific manner by ICAM-1, ICAM-3 and VCAM-1 and can be inhibited by antibodies to integrin beta 2 or beta 7. J. Gen. Virol. 80:1429-1436. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

