Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids
- PMID: 12941896
- PMCID: PMC224603
- DOI: 10.1128/jvi.77.18.9862-9871.2003
Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids
Abstract
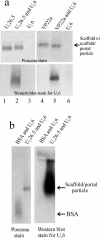
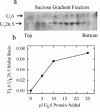
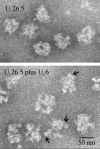
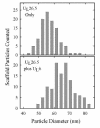
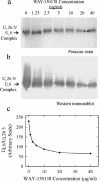
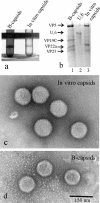
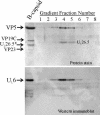
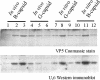
The herpes simplex virus type 1 (HSV-1) portal complex is a ring-shaped structure located at a single vertex in the viral capsid. Composed of 12 U(L)6 protein molecules, the portal functions as a channel through which DNA passes as it enters the capsid. The studies described here were undertaken to clarify how the portal becomes incorporated as the capsid is assembled. We tested the idea that an intact portal may be donated to the growing capsid by way of a complex with the major scaffolding protein, U(L)26.5. Soluble U(L)26.5-portal complexes were found to assemble when purified portals were mixed in vitro with U(L)26.5. The complexes, called scaffold-portal particles, were stable during purification by agarose gel electrophoresis or sucrose density gradient ultracentrifugation. Examination of the scaffold-portal particles by electron microscopy showed that they resemble the 50- to 60-nm-diameter "scaffold particles" formed from purified U(L)26.5. They differed, however, in that intact portals were observed on the surface. Analysis of the protein composition by sodium dodecyl sulfate-polyacrylamide gel electrophoresis demonstrated that portals and U(L)26.5 combine in various proportions, with the highest observed U(L)6 content corresponding to two or three portals per scaffold particle. Association between the portal and U(L)26.5 was antagonized by WAY-150138, a small-molecule inhibitor of HSV-1 replication. Soluble scaffold-portal particles were found to function in an in vitro capsid assembly system that also contained the major capsid (VP5) and triplex (VP19C and VP23) proteins. Capsids that formed in this system had the structure and protein composition expected of mature HSV-1 capsids, including U(L)6, at a level corresponding to approximately 1 portal complex per capsid. The results support the view that U(L)6 becomes incorporated into nascent HSV-1 capsids by way of a complex with U(L)26.5 and suggest further that U(L)6 may be introduced into the growing capsid as an intact portal.
Figures
Similar articles
-
Herpesvirus capsid assembly: insights from structural analysis.Curr Opin Virol. 2011 Aug;1(2):142-9. doi: 10.1016/j.coviro.2011.06.003. Curr Opin Virol. 2011. PMID: 21927635 Free PMC article. Review.
-
Involvement of the portal at an early step in herpes simplex virus capsid assembly.J Virol. 2005 Aug;79(16):10540-6. doi: 10.1128/JVI.79.16.10540-10546.2005. J Virol. 2005. PMID: 16051846 Free PMC article.
-
Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation.J Mol Biol. 1996 Nov 1;263(3):432-46. doi: 10.1006/jmbi.1996.0587. J Mol Biol. 1996. PMID: 8918599
-
The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly.J Mol Biol. 1996 Nov 1;263(3):447-62. doi: 10.1016/s0022-2836(96)80018-0. J Mol Biol. 1996. PMID: 8918600
-
Herpesvirus Capsid Assembly and DNA Packaging.Adv Anat Embryol Cell Biol. 2017;223:119-142. doi: 10.1007/978-3-319-53168-7_6. Adv Anat Embryol Cell Biol. 2017. PMID: 28528442 Free PMC article. Review.
Cited by
-
Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids.J Virol. 2006 Mar;80(5):2118-26. doi: 10.1128/JVI.80.5.2118-2126.2006. J Virol. 2006. PMID: 16474120 Free PMC article.
-
Alpha-herpesvirus infection induces the formation of nuclear actin filaments.PLoS Pathog. 2006 Aug;2(8):e85. doi: 10.1371/journal.ppat.0020085. PLoS Pathog. 2006. PMID: 16933992 Free PMC article.
-
Herpesvirus capsid assembly: insights from structural analysis.Curr Opin Virol. 2011 Aug;1(2):142-9. doi: 10.1016/j.coviro.2011.06.003. Curr Opin Virol. 2011. PMID: 21927635 Free PMC article. Review.
-
Disulfide bond formation in the herpes simplex virus 1 UL6 protein is required for portal ring formation and genome encapsidation.J Virol. 2011 Sep;85(17):8616-24. doi: 10.1128/JVI.00123-11. Epub 2011 May 18. J Virol. 2011. PMID: 21593161 Free PMC article.
-
Nuclear localization sequences in cytomegalovirus capsid assembly proteins (UL80 proteins) are required for virus production: inactivating NLS1, NLS2, or both affects replication to strikingly different extents.J Virol. 2008 Jun;82(11):5381-9. doi: 10.1128/JVI.02697-07. Epub 2008 Mar 19. J Virol. 2008. PMID: 18353959 Free PMC article.
References
-
- Bazinet, C., J. Benbasat, J. King, J. M. Carazo, and J. L. Carrascosa. 1988. Purification and organization of the gene 1 portal protein required for phage P22 DNA packaging. Biochemistry 27:1849-1856. - PubMed
-
- Bazinet, C., and J. King. 1988. Initiation of P22 procapsid assembly in vivo. J. Mol. Biol. 202:77-86. - PubMed
-
- Brown, J. C., M. A. McVoy, and F. L. Homa. 2001. Packaging DNA into herpesvirus capsids, p. 111-153. In A. Holzenburg and E. Bogner (ed.), Structure-function relationships of human pathogenic viruses. Kluwer Academic Publishers, London, United Kingdom.
-
- Eiserling, F. A., and L. W. Black. 1994. Pathways in T4 morphogenesis, p. 209-212. In J. D. Karam (ed.), Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
-
- Engvall, E., and P. Perlman. 1971. Enzyme-linked immunosorbent assay (ELISA): quantitative assay for immunoglobulin. Immunochemistry 8:871-874. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

