Human papillomavirus type 16 E6 activates TERT gene transcription through induction of c-Myc and release of USF-mediated repression
- PMID: 12941894
- PMCID: PMC224601
- DOI: 10.1128/jvi.77.18.9852-9861.2003
Human papillomavirus type 16 E6 activates TERT gene transcription through induction of c-Myc and release of USF-mediated repression
Abstract
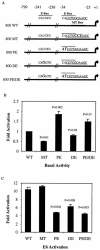
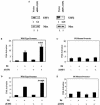
Human papillomavirus type 16 (HPV-16), a DNA tumor virus, has a causal role in cervical cancer, and the viral oncoproteins E6 and E7 contribute to oncogenesis in multiple ways. E6 increases telomerase activity in keratinocytes through increased transcription of the telomerase catalytic subunit gene (TERT), but the factors involved in this have been elusive. We have found that mutation of the proximal E box in the TERT promoter has an activating effect in luciferase assays. This suggested that a repressive complex might be present at this site. HPV-16 E6 activated the TERT promoter predominantly through the proximal E box, and thus, might be acting on this repressive complex. This site is specific for the Myc/Mad/Max transcription factors as well as USF1 and USF2. Addition of exogenous USF1 or USF2 repressed activation of the TERT promoter by E6, dependent on the proximal E box. Using siRNA against USF1 or USF2 allowed for greater activation of the TERT promoter by E6. Conversely, loss of c-Myc function, through a dominant-negative Myc molecule, reduced activation by E6. Chromatin immunoprecipitations showed that in the presence of E6, there was a reduction in binding of USF1 and USF2 at the TERT promoter proximal E box, and a concomitant increase in c-Myc bound to this site. This shows that a repressive complex containing USF1 and USF2 is present in normal cells with little or no telomerase activity. In E6 keratinocytes, this repressive complex is replaced by c-Myc, which corresponds to higher levels of TERT transcription and consequently, telomerase activity.
Figures
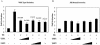

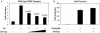

Similar articles
-
Human papillomavirus E6 and Myc proteins associate in vivo and bind to and cooperatively activate the telomerase reverse transcriptase promoter.Proc Natl Acad Sci U S A. 2003 Jul 8;100(14):8211-6. doi: 10.1073/pnas.1435900100. Epub 2003 Jun 23. Proc Natl Acad Sci U S A. 2003. PMID: 12821782 Free PMC article.
-
Telomerase activation by human papillomavirus type 16 E6 protein: induction of human telomerase reverse transcriptase expression through Myc and GC-rich Sp1 binding sites.J Virol. 2001 Jun;75(12):5559-66. doi: 10.1128/JVI.75.12.5559-5566.2001. J Virol. 2001. PMID: 11356963 Free PMC article.
-
Cell-restricted immortalization by human papillomavirus correlates with telomerase activation and engagement of the hTERT promoter by Myc.J Virol. 2008 Dec;82(23):11568-76. doi: 10.1128/JVI.01318-08. Epub 2008 Sep 25. J Virol. 2008. PMID: 18818322 Free PMC article.
-
Transcriptional Regulation of Telomerase Reverse Transcriptase (TERT) by MYC.Front Cell Dev Biol. 2017 Jan 26;5:1. doi: 10.3389/fcell.2017.00001. eCollection 2017. Front Cell Dev Biol. 2017. PMID: 28184371 Free PMC article. Review.
-
The Molecular Interplay between Human Oncoviruses and Telomerase in Cancer Development.Cancers (Basel). 2022 Oct 26;14(21):5257. doi: 10.3390/cancers14215257. Cancers (Basel). 2022. PMID: 36358677 Free PMC article. Review.
Cited by
-
Telomerase as a useful target in cancer fighting-the breast cancer case.Tumour Biol. 2013 Jun;34(3):1371-80. doi: 10.1007/s13277-013-0757-4. Epub 2013 Apr 5. Tumour Biol. 2013. PMID: 23558965 Free PMC article. Review.
-
Papillomavirus E6 oncoproteins.Virology. 2013 Oct;445(1-2):115-37. doi: 10.1016/j.virol.2013.04.026. Epub 2013 May 24. Virology. 2013. PMID: 23711382 Free PMC article. Review.
-
Decreased migration of Langerhans precursor-like cells in response to human keratinocytes expressing human papillomavirus type 16 E6/E7 is related to reduced macrophage inflammatory protein-3alpha production.J Virol. 2005 Dec;79(23):14852-62. doi: 10.1128/JVI.79.23.14852-14862.2005. J Virol. 2005. PMID: 16282485 Free PMC article.
-
High-Risk Human Papillomaviral Oncogenes E6 and E7 Target Key Cellular Pathways to Achieve Oncogenesis.Int J Mol Sci. 2018 Jun 8;19(6):1706. doi: 10.3390/ijms19061706. Int J Mol Sci. 2018. PMID: 29890655 Free PMC article. Review.
-
Epidermal growth factor activates telomerase activity by direct binding of Ets-2 to hTERT promoter in lung cancer cells.Tumour Biol. 2015 Jul;36(7):5389-98. doi: 10.1007/s13277-015-3204-x. Epub 2015 Feb 14. Tumour Biol. 2015. PMID: 25680408
References
-
- Amati, B., M. W. Brooks, N. Levy, T. D. Littlewood, G. I. Evan, and H. Land. 1993. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell 72:233-245. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

