The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum
- PMID: 12929205
- PMCID: PMC176545
- DOI: 10.1371/journal.pbio.0000005
The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum
Abstract
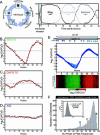
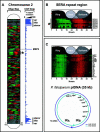
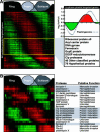
Plasmodium falciparum is the causative agent of the most burdensome form of human malaria, affecting 200-300 million individuals per year worldwide. The recently sequenced genome of P. falciparum revealed over 5,400 genes, of which 60% encode proteins of unknown function. Insights into the biochemical function and regulation of these genes will provide the foundation for future drug and vaccine development efforts toward eradication of this disease. By analyzing the complete asexual intraerythrocytic developmental cycle (IDC) transcriptome of the HB3 strain of P. falciparum, we demonstrate that at least 60% of the genome is transcriptionally active during this stage. Our data demonstrate that this parasite has evolved an extremely specialized mode of transcriptional regulation that produces a continuous cascade of gene expression, beginning with genes corresponding to general cellular processes, such as protein synthesis, and ending with Plasmodium-specific functionalities, such as genes involved in erythrocyte invasion. The data reveal that genes contiguous along the chromosomes are rarely coregulated, while transcription from the plastid genome is highly coregulated and likely polycistronic. Comparative genomic hybridization between HB3 and the reference genome strain (3D7) was used to distinguish between genes not expressed during the IDC and genes not detected because of possible sequence variations. Genomic differences between these strains were found almost exclusively in the highly antigenic subtelomeric regions of chromosomes. The simple cascade of gene regulation that directs the asexual development of P. falciparum is unprecedented in eukaryotic biology. The transcriptome of the IDC resembles a "just-in-time" manufacturing process whereby induction of any given gene occurs once per cycle and only at a time when it is required. These data provide to our knowledge the first comprehensive view of the timing of transcription throughout the intraerythrocytic development of P. falciparum and provide a resource for the identification of new chemotherapeutic and vaccine candidates.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures


Comment in
-
Opening access to cell biology.PLoS Biol. 2005 Dec;3(12):e426. doi: 10.1371/journal.pbio.0030426. PLoS Biol. 2005. PMID: 17593892 Free PMC article.
-
A rising tide of parasite transcriptomics propels pathogen biology.PLoS Biol. 2023 Jan 25;21(1):e3001997. doi: 10.1371/journal.pbio.3001997. eCollection 2023 Jan. PLoS Biol. 2023. PMID: 36696650 Free PMC article.
Similar articles
-
A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs.Genome Biol. 2011 Jun 20;12(6):R56. doi: 10.1186/gb-2011-12-6-r56. Genome Biol. 2011. PMID: 21689454 Free PMC article.
-
Genome-wide real-time in vivo transcriptional dynamics during Plasmodium falciparum blood-stage development.Nat Commun. 2018 Jul 9;9(1):2656. doi: 10.1038/s41467-018-04966-3. Nat Commun. 2018. PMID: 29985403 Free PMC article.
-
Patterns of gene-specific and total transcriptional activity during the Plasmodium falciparum intraerythrocytic developmental cycle.Eukaryot Cell. 2009 Mar;8(3):327-38. doi: 10.1128/EC.00340-08. Epub 2009 Jan 16. Eukaryot Cell. 2009. PMID: 19151330 Free PMC article.
-
Pernicious plans revealed: Plasmodium falciparum genome wide expression analysis.Curr Opin Microbiol. 2004 Aug;7(4):382-7. doi: 10.1016/j.mib.2004.06.014. Curr Opin Microbiol. 2004. PMID: 15358256 Review.
-
Functional genomics, new tools in malaria research.Ann Ist Super Sanita. 2005;41(4):469-77. Ann Ist Super Sanita. 2005. PMID: 16569915 Review.
Cited by
-
Neurotransmitters and molecular chaperones interactions in cerebral malaria: Is there a missing link?Front Mol Biosci. 2022 Aug 24;9:965569. doi: 10.3389/fmolb.2022.965569. eCollection 2022. Front Mol Biosci. 2022. PMID: 36090033 Free PMC article. Review.
-
Plasmodium RON11 triggers biogenesis of the merozoite rhoptry pair and is essential for erythrocyte invasion.PLoS Biol. 2024 Sep 18;22(9):e3002801. doi: 10.1371/journal.pbio.3002801. eCollection 2024 Sep. PLoS Biol. 2024. PMID: 39292724 Free PMC article.
-
A Proteasome Mutation Sensitizes P. falciparum Cam3.II K13C580Y Parasites to DHA and OZ439.ACS Infect Dis. 2021 Jul 9;7(7):1923-1931. doi: 10.1021/acsinfecdis.0c00900. Epub 2021 May 10. ACS Infect Dis. 2021. PMID: 33971094 Free PMC article.
-
Variation in selective constraints along the Plasmodium life cycle.Infect Genet Evol. 2021 Aug;92:104908. doi: 10.1016/j.meegid.2021.104908. Epub 2021 May 8. Infect Genet Evol. 2021. PMID: 33975022 Free PMC article.
-
Differential Gene Expression of Malaria Parasite in Response to Red Blood Cell-Specific Glycolytic Intermediate 2,3-Diphosphoglycerate (2,3-DPG).Int J Mol Sci. 2023 Nov 28;24(23):16869. doi: 10.3390/ijms242316869. Int J Mol Sci. 2023. PMID: 38069204 Free PMC article.
References
-
- Afonso Nogueira P, Wunderlich G, Shugiro Tada M, d'Arc Neves Costa J, Jose Menezeset M, et al. Plasmodium falciparum: Analysis of transcribed var gene sequences in natural isolates from the Brazilian Amazon region. Exp Parasitol. 2002;101:111–120. - PubMed
-
- Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, et al. Gene expression during the life cycle of Drosophila melanogaster . Science. 2002;297:2270–2275. - PubMed
-
- Ben Mamoun C, Gluzman IY, Hott C, MacMillan SK, Amarakone AS, et al. Co-ordinated programme of gene expression during asexual intraerythrocytic development of the human malaria parasite Plasmodium falciparum revealed by microarray analysis. Mol Microbiol. 2001;39:26–36. - PubMed
-
- Blackman MJ. Proteases involved in erythrocyte invasion by the malaria parasite: Function and potential as chemotherapeutic targets. Curr Drug Targets. 2000;1:59–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

