Automated extraction and quantification of human cytomegalovirus DNA in whole blood by real-time PCR assay
- PMID: 12904398
- PMCID: PMC179853
- DOI: 10.1128/JCM.41.8.3840-3845.2003
Automated extraction and quantification of human cytomegalovirus DNA in whole blood by real-time PCR assay
Abstract
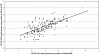
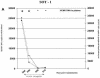
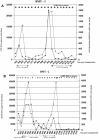
The measurement of human cytomegalovirus (HCMV) DNA in blood is becoming the standard method for monitoring HCMV infection in immune-suppressed and unsuppressed patients. As various blood compartments can be used, we have compared the HCMV DNA measured in whole blood (WB), peripheral blood leukocytes (PBL), and plasma by real-time PCR. We tested 286 samples: HCMV DNA was extracted automatically from WB and PBL with the MagNA Pure instrument (Roche Molecular Biochemicals) and manually from plasma samples. The HCMV DNA from WB, PBL, and plasma was measured by real-time Light Cycler PCR. Primers and probe were located in the UL 83 region. HCMV DNA was detected more frequently in WB (88.5%) than in the PBL (65.7%) (P < 0.0001) or the plasma (55.2%) (P < 0.0001). There was a good correlation between the positive results in WB and in PBL (r = 0.68; P < 0.0001), and 3.15 log(10) genome copies in 200000 PBL, equivalent to the threshold value of 50 pp65-positive polymorphonuclear cells per 200000 leukocytes, was equivalent to 3.4 log(10) genome copies in 200 microl of WB. WB was shown to be suitable for automated extraction and the quantitation of HCMV DNA by real-time Light Cycler PCR by analysis of serial samples from representative patients of various populations. This system may be very useful for monitoring of immune-suppressed and unsuppressed patients.
Figures




Similar articles
-
Quantitation of human cytomegalovirus in recipients of solid organ transplants by real-time quantitative PCR and pp65 antigenemia.J Med Virol. 2003 Feb;69(2):225-31. doi: 10.1002/jmv.10277. J Med Virol. 2003. PMID: 12683412
-
Quantitative analysis of HCMV DNA load in whole blood of renal transplant patients using real-time PCR assay.J Clin Virol. 2004 Mar;29(3):194-201. doi: 10.1016/S1386-6532(03)00124-0. J Clin Virol. 2004. PMID: 14962789
-
Monitoring of human cytomegalovirus infection in immunosuppressed patients using real-time PCR on whole blood.J Clin Virol. 2007 Nov;40(3):173-9. doi: 10.1016/j.jcv.2007.08.014. Epub 2007 Sep 29. J Clin Virol. 2007. PMID: 17904901
-
Whole blood real-time quantitative PCR for cytomegalovirus infection follow-up in transplant recipients.J Clin Virol. 2006 May;36(1):72-5. doi: 10.1016/j.jcv.2006.01.002. Epub 2006 Feb 14. J Clin Virol. 2006. PMID: 16481215
-
Use of the human cytomegalovirus (HCMV) antigenemia assay for diagnosis and monitoring of HCMV infections and detection of antiviral drug resistance in the immunocompromised.J Clin Virol. 1998 Jul 24;11(1):51-60. doi: 10.1016/s0928-0197(98)00040-3. J Clin Virol. 1998. PMID: 9784143 Review.
Cited by
-
Comparative evaluation of a commercially available automated system for extraction of viral DNA from whole blood: application to monitoring of epstein-barr virus and cytomegalovirus load.J Clin Microbiol. 2009 Nov;47(11):3753-5. doi: 10.1128/JCM.01497-09. Epub 2009 Aug 26. J Clin Microbiol. 2009. PMID: 19710270 Free PMC article.
-
An Association between BK Virus Replication in Bone Marrow and Cytopenia in Kidney-Transplant Recipients.J Transplant. 2014;2014:252914. doi: 10.1155/2014/252914. Epub 2014 Apr 29. J Transplant. 2014. PMID: 24868448 Free PMC article.
-
Human cytomegalovirus UL23 exploits PD-L1 inhibitory signaling pathway to evade T cell-mediated cytotoxicity.mBio. 2024 Jul 17;15(7):e0119124. doi: 10.1128/mbio.01191-24. Epub 2024 Jun 3. mBio. 2024. PMID: 38829126 Free PMC article.
-
Rapid quantitation of cytomegalovirus DNA in whole blood by a new molecular assay based on automated sample preparation and real-time PCR.Med Microbiol Immunol. 2010 Nov;199(4):311-6. doi: 10.1007/s00430-010-0164-z. Epub 2010 Jun 18. Med Microbiol Immunol. 2010. PMID: 20559848
-
Comparison of five methods for extraction of Legionella pneumophila from respiratory specimens.J Clin Microbiol. 2004 Dec;42(12):5913-6. doi: 10.1128/JCM.42.12.5913-5916.2004. J Clin Microbiol. 2004. PMID: 15583339 Free PMC article.
References
-
- Boivin, G., R. Belanger, R. Delage, C. Beliveau, C. Demers, N. Goyette, and J. Roy. 2000. Quantitative analysis of cytomegalovirus (CMV) viremia using the pp65 antigenemia assay and the COBAS AMPLICOR CMV MONITOR PCR test after blood and marrow allogeneic transplantation. J. Clin. Microbiol. 38:4356-4360. - PMC - PubMed
-
- Boivin, G., J. Handfield, E. Toma, G. Murray, R. Lalonde, and M. G. Bergeron. 1998. Comparative evaluation of the cytomegalovirus DNA load in polymorphonuclear leukocytes and plasma of human immunodeficiency virus-infected subjects. J. Infect. Dis. 177:355-360. - PubMed
-
- Freymuth, F., E. Gennetay, J. Petitjean, G. Eugene, B. Hurault de Ligny, J. P. Ryckelynck, C. Legoff, P. Hazera, and C. Bazin. 1994. Comparison of nested PCR for detection of DNA in plasma with pp65 leukocytic antigenemia procedure for diagnosis of human cytomegalovirus infection. J. Clin. Microbiol. 32:1614-1618. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

