HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein
- PMID: 12900394
- PMCID: PMC2172688
- DOI: 10.1083/jcb.200302138
HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein
Abstract
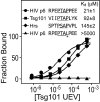
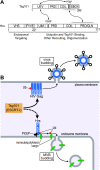
The HIV-1 Gag protein recruits the cellular factor Tsg101 to facilitate the final stages of virus budding. A conserved P(S/T)AP tetrapeptide motif within Gag (the "late domain") binds directly to the NH2-terminal ubiquitin E2 variant (UEV) domain of Tsg101. In the cell, Tsg101 is required for biogenesis of vesicles that bud into the lumen of late endosomal compartments called multivesicular bodies (MVBs). However, the mechanism by which Tsg101 is recruited from the cytoplasm onto the endosomal membrane has not been known. Now, we report that Tsg101 binds the COOH-terminal region of the endosomal protein hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs; residues 222-777). This interaction is mediated, in part, by binding of the Tsg101 UEV domain to the Hrs 348PSAP351 motif. Importantly, Hrs222-777 can recruit Tsg101 and rescue the budding of virus-like Gag particles that are missing native late domains. These observations indicate that Hrs normally functions to recruit Tsg101 to the endosomal membrane. HIV-1 Gag apparently mimics this Hrs activity, and thereby usurps Tsg101 and other components of the MVB vesicle fission machinery to facilitate viral budding.
Figures





Similar articles
-
After Hrs with HIV.J Cell Biol. 2003 Aug 4;162(3):371-5. doi: 10.1083/jcb.200307062. J Cell Biol. 2003. PMID: 12900390 Free PMC article. Review.
-
Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes.J Cell Biol. 2003 Aug 4;162(3):435-42. doi: 10.1083/jcb.200302131. J Cell Biol. 2003. PMID: 12900395 Free PMC article.
-
HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress.Nat Med. 2001 Dec;7(12):1313-9. doi: 10.1038/nm1201-1313. Nat Med. 2001. PMID: 11726971
-
Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding.J Cell Sci. 2004 May 1;117(Pt 11):2357-67. doi: 10.1242/jcs.01095. J Cell Sci. 2004. PMID: 15126635
-
[HIV budding and Tsg101].Uirusu. 2005 Dec;55(2):281-6. doi: 10.2222/jsv.55.281. Uirusu. 2005. PMID: 16557014 Review. Japanese.
Cited by
-
Biogenesis of Extracellular Vesicles.Subcell Biochem. 2021;97:19-43. doi: 10.1007/978-3-030-67171-6_2. Subcell Biochem. 2021. PMID: 33779912
-
Exosomes: Implications in HIV-1 Pathogenesis.Viruses. 2015 Jul 20;7(7):4093-118. doi: 10.3390/v7072810. Viruses. 2015. PMID: 26205405 Free PMC article. Review.
-
Tsg101/ESCRT-I recruitment regulated by the dual binding modes of K63-linked diubiquitin.Structure. 2022 Feb 3;30(2):289-299.e6. doi: 10.1016/j.str.2021.09.006. Structure. 2022. PMID: 35120596 Free PMC article.
-
The ESCRT complexes: structure and mechanism of a membrane-trafficking network.Annu Rev Biophys Biomol Struct. 2006;35:277-98. doi: 10.1146/annurev.biophys.35.040405.102126. Annu Rev Biophys Biomol Struct. 2006. PMID: 16689637 Free PMC article. Review.
-
The cell biology of HIV-1 virion genesis.Cell Host Microbe. 2009 Jun 18;5(6):550-8. doi: 10.1016/j.chom.2009.05.015. Cell Host Microbe. 2009. PMID: 19527882 Free PMC article. Review.
References
-
- Asao, H., Y. Sasaki, T. Arita, N. Tanaka, K. Endo, H. Kasai, T. Takeshita, Y. Endo, T. Fujita, and K. Sugamura. 1997. Hrs is associated with STAM, a signal-transducing adaptor molecule. Its suppressive effect on cytokine-induced cell growth. J. Biol. Chem. 272:32785–32791. - PubMed
-
- Babst, M., D. Katzmann, E. Estepa-Sabal, T. Meerloo, and S. Emr. 2002. a. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 3:271–282. - PubMed
-
- Babst, M., D. Katzmann, W. Snyder, B. Wendland, and S. Emr. 2002. b. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 3:283–289. - PubMed
-
- Bache, K.G., C. Raiborg, A. Mehlum, I.H. Madshus, and H. Stenmark. 2002. Phosphorylation of Hrs downstream of the epidermal growth factor receptor. Eur. J. Biochem. 269:3881–3887. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

