Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes
- PMID: 12881432
- PMCID: PMC169057
- DOI: 10.1093/emboj/cdg384
Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes
Abstract
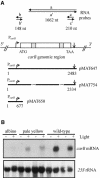
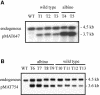
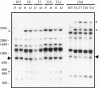
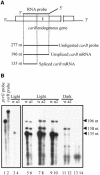
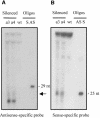
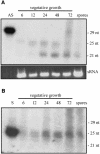
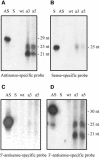
Transformation of Mucor circinelloides with self-replicative plasmids containing a wild-type copy of the carotenogenic gene carB causes silencing of the carB function in 3% of transformants. Genomic analyses revealed a relationship between silenced phenotype and number of copies of plasmids. This phenotype results from a reduction of the steady-state levels of carB mRNA, a reduction that is not due to differences in the level of transcription, indicating that silencing is post-transcriptional. Small sense and antisense RNAs have been found to be associated with gene silencing in M. circinelloides. Two size classes of small antisense RNAs, differentially accumulated during the vegetative growth of silenced transformants, have been detected: a long 25-nucleotide RNA and a short 21-nucleotide RNA. Secondary sense and antisense RNAs corresponding to sequences of the endogenous gene downstream of the initial triggering molecule have also been detected, revealing the existence of spreading of RNA targeting in fungi. These findings, together with the self-replicative nature of the triggering molecules, make M. circinelloides a suitable organism for investigating some unresolved questions in RNA silencing.
Figures
Similar articles
-
Two distinct RNA-dependent RNA polymerases are required for initiation and amplification of RNA silencing in the basal fungus Mucor circinelloides.Mol Microbiol. 2012 Jan;83(2):379-94. doi: 10.1111/j.1365-2958.2011.07939.x. Epub 2011 Dec 19. Mol Microbiol. 2012. PMID: 22141923
-
A single dicer gene is required for efficient gene silencing associated with two classes of small antisense RNAs in Mucor circinelloides.Eukaryot Cell. 2009 Oct;8(10):1486-97. doi: 10.1128/EC.00191-09. Epub 2009 Aug 7. Eukaryot Cell. 2009. PMID: 19666782 Free PMC article.
-
Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects.Fungal Genet Biol. 2007 Jun;44(6):504-16. doi: 10.1016/j.fgb.2006.09.003. Epub 2006 Oct 30. Fungal Genet Biol. 2007. PMID: 17074518
-
Quelling: post-transcriptional gene silencing guided by small RNAs in Neurospora crassa.Curr Opin Microbiol. 2007 Apr;10(2):199-203. doi: 10.1016/j.mib.2007.03.016. Epub 2007 Mar 28. Curr Opin Microbiol. 2007. PMID: 17395524 Review.
-
RNAi pathways in Mucor: A tale of proteins, small RNAs and functional diversity.Fungal Genet Biol. 2016 May;90:44-52. doi: 10.1016/j.fgb.2015.11.006. Epub 2015 Nov 21. Fungal Genet Biol. 2016. PMID: 26593631 Review.
Cited by
-
The Whys and Wherefores of Transitivity in Plants.Front Plant Sci. 2020 Aug 31;11:579376. doi: 10.3389/fpls.2020.579376. eCollection 2020. Front Plant Sci. 2020. PMID: 32983223 Free PMC article. Review.
-
A non-canonical RNAi pathway controls virulence and genome stability in Mucorales.PLoS Genet. 2020 Jul 13;16(7):e1008611. doi: 10.1371/journal.pgen.1008611. eCollection 2020 Jul. PLoS Genet. 2020. PMID: 32658892 Free PMC article.
-
Evolution and diversification of RNA silencing proteins in fungi.J Mol Evol. 2006 Jul;63(1):127-35. doi: 10.1007/s00239-005-0257-2. Epub 2006 Jun 16. J Mol Evol. 2006. PMID: 16786437
-
Stable and reproducible homologous recombination enables CRISPR-based engineering in the fungus Rhizopus microsporus.Cell Rep Methods. 2021 Dec 6;1(8):100124. doi: 10.1016/j.crmeth.2021.100124. eCollection 2021 Dec 20. Cell Rep Methods. 2021. PMID: 35475217 Free PMC article.
-
The RNAi machinery controls distinct responses to environmental signals in the basal fungus Mucor circinelloides.BMC Genomics. 2015 Mar 25;16(1):237. doi: 10.1186/s12864-015-1443-2. BMC Genomics. 2015. PMID: 25880254 Free PMC article.
References
-
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1989) Current Protocols in Molecular Biology. Green Publishing Associates and Wiley-Interscience, New York, NY.
-
- Benito E.P., Campuzano,V., López-Matas,M.A., De Vicente,J.I. and Eslava,A.P. (1995) Isolation, characterization and transformation, by autonomous replication, of Mucor circinelloides OMPdecase-deficient mutants. Mol. Gen. Genet., 248, 126–135. - PubMed
-
- Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

