Protein transport in plant cells: in and out of the Golgi
- PMID: 12876187
- PMCID: PMC4243656
- DOI: 10.1093/aob/mcg134
Protein transport in plant cells: in and out of the Golgi
Erratum in
- Ann Bot (Lond). 2003 Sep;92(3):475
Abstract
In plant cells, the Golgi apparatus is the key organelle for polysaccharide and glycolipid synthesis, protein glycosylation and protein sorting towards various cellular compartments. Protein import from the endoplasmic reticulum (ER) is a highly dynamic process, and new data suggest that transport, at least of soluble proteins, occurs via bulk flow. In this Botanical Briefing, we review the latest data on ER/Golgi inter-relations and the models for transport between the two organelles. Whether vesicles are involved in this transport event or if direct ER-Golgi connections exist are questions that are open to discussion. Whereas the majority of proteins pass through the Golgi on their way to other cell destinations, either by vesicular shuttles or through maturation of cisternae from the cis- to the trans-face, a number of membrane proteins reside in the different Golgi cisternae. Experimental evidence suggests that the length of the transmembrane domain is of crucial importance for the retention of proteins within the Golgi. In non-dividing cells, protein transport out of the Golgi is either directed towards the plasma membrane/cell wall (secretion) or to the vacuolar system. The latter comprises the lytic vacuole and protein storage vacuoles. In general, transport to either of these from the Golgi depends on different sorting signals and receptors and is mediated by clathrin-coated and dense vesicles, respectively. Being at the heart of the secretory pathway, the Golgi (transiently) accommodates regulatory proteins of secretion (e.g. SNAREs and small GTPases), of which many have been cloned in plants over the last decade. In this context, we present a list of regulatory proteins, along with structural and processing proteins, that have been located to the Golgi and the 'trans-Golgi network' by microscopy.
Figures
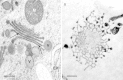
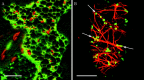
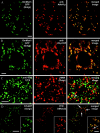
Similar articles
-
Vesicles versus Tubes: Is Endoplasmic Reticulum-Golgi Transport in Plants Fundamentally Different from Other Eukaryotes?Plant Physiol. 2015 Jun;168(2):393-406. doi: 10.1104/pp.15.00124. Epub 2015 Apr 16. Plant Physiol. 2015. PMID: 25883241 Free PMC article. Review.
-
A SNARE involved in protein transport through the Golgi apparatus.Nature. 1997 Oct 23;389(6653):881-4. doi: 10.1038/39923. Nature. 1997. PMID: 9349823
-
Unconventional protein secretion.Trends Plant Sci. 2012 Oct;17(10):606-15. doi: 10.1016/j.tplants.2012.06.004. Epub 2012 Jul 9. Trends Plant Sci. 2012. PMID: 22784825 Review.
-
Plant Golgi ultrastructure.J Microsc. 2020 Nov;280(2):111-121. doi: 10.1111/jmi.12899. Epub 2020 May 27. J Microsc. 2020. PMID: 32420623 Review.
-
Formation and maintenance of the Golgi apparatus in plant cells.Int Rev Cell Mol Biol. 2014;310:221-87. doi: 10.1016/B978-0-12-800180-6.00006-2. Int Rev Cell Mol Biol. 2014. PMID: 24725428 Review.
Cited by
-
Differential effects of human and plant N-acetylglucosaminyltransferase I (GnTI) in plants.Transgenic Res. 2010 Aug;19(4):535-47. doi: 10.1007/s11248-009-9331-7. Epub 2009 Oct 14. Transgenic Res. 2010. PMID: 19826906 Free PMC article.
-
Clathrin is involved in organization of mitotic spindle and phragmoplast as well as in endocytosis in tobacco cell cultures.Protoplasma. 2007;230(1-2):1-11. doi: 10.1007/s00709-006-0226-7. Epub 2007 Mar 13. Protoplasma. 2007. PMID: 17351731
-
Genome-Wide Association Study of Seed Morphology Traits in Senegalese Sorghum Cultivars.Plants (Basel). 2023 Jun 16;12(12):2344. doi: 10.3390/plants12122344. Plants (Basel). 2023. PMID: 37375969 Free PMC article.
-
Predicting protein subcellular localization: past, present, and future.Genomics Proteomics Bioinformatics. 2004 Nov;2(4):209-15. doi: 10.1016/s1672-0229(04)02027-3. Genomics Proteomics Bioinformatics. 2004. PMID: 15901249 Free PMC article. Review.
-
Increasing the Efficiency of the Accumulation of Recombinant Proteins in Plant Cells: The Role of Transport Signal Peptides.Plants (Basel). 2022 Sep 28;11(19):2561. doi: 10.3390/plants11192561. Plants (Basel). 2022. PMID: 36235427 Free PMC article. Review.
References
-
- AllanBB, Moyer BD, Balch WE.2000. Rab1 recruitment of p115 into a cis‐SNARE complex: programming budding COPII vesicles for fusion. Science 289: 444–448. - PubMed
-
- AndreevaAV, Kutuzov MA, Evans DE, Hawes CR.1998a. Proteins involved in membrane transport between the ER and the Golgi apparatus: 21 putative plant homologues revealed by dbEST searching. Cell Biology International 22: 145–160. - PubMed
-
- AndreevaAV, Kutuzov MA, Evans DE, Hawes CR.1998b. The structure and function of the Golgi apparatus: a hundred years of questions. Journal of Experimental Botany 49: 1281–1291.
-
- AndreevaAV, Zheng H, Saint‐Jore CM, Kutuzov MA, Evans DE, Hawes CR.2000. Organization of transport from endoplasmic reticulum to Golgi in higher plants. Biochemical Society Transactions 28: 505–512. - PubMed
-
- BarloweC.2002. COPII‐dependent transport from the endoplasmic reticulum. Current Opinion in Cell Biology 14: 417–422. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

