Structure of a trapped endonuclease III-DNA covalent intermediate
- PMID: 12840008
- PMCID: PMC165637
- DOI: 10.1093/emboj/cdg311
Structure of a trapped endonuclease III-DNA covalent intermediate
Abstract
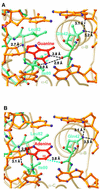
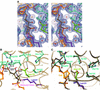
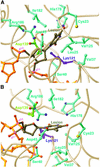
Nearly all cells express proteins that confer resistance to the mutagenic effects of oxidative DNA damage. The primary defense against the toxicity of oxidative nucleobase lesions in DNA is the base-excision repair (BER) pathway. Endonuclease III (EndoIII) is a [4Fe-4S] cluster-containing DNA glycosylase with repair activity specific for oxidized pyrimidine lesions in duplex DNA. We have determined the crystal structure of a trapped intermediate that represents EndoIII frozen in the act of repairing DNA. The structure of the protein-DNA complex provides insight into the ability of EndoIII to recognize and repair a diverse array of oxidatively damaged bases. This structure also suggests a rationale for the frequent occurrence in certain human cancers of a specific mutation in the related DNA repair protein MYH.
Figures

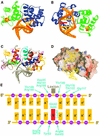
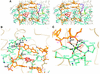
Similar articles
-
Structure and function of Escherichia coli endonuclease III.Ann N Y Acad Sci. 1994 Jul 29;726:215-22. doi: 10.1111/j.1749-6632.1994.tb52818.x. Ann N Y Acad Sci. 1994. PMID: 8092678 Review. No abstract available.
-
Three-dimensional structural views of damaged-DNA recognition: T4 endonuclease V, E. coli Vsr protein, and human nucleotide excision repair factor XPA.Mutat Res. 2000 Aug 30;460(3-4):257-75. doi: 10.1016/s0921-8777(00)00031-8. Mutat Res. 2000. PMID: 10946233 Review.
-
Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition.Cell. 1995 Dec 1;83(5):773-82. doi: 10.1016/0092-8674(95)90190-6. Cell. 1995. PMID: 8521494
-
The role of the iron-sulfur cluster in Escherichia coli endonuclease III. A resonance Raman study.J Biol Chem. 1992 Aug 15;267(23):16135-7. J Biol Chem. 1992. PMID: 1644800
-
Atomic structure of the DNA repair [4Fe-4S] enzyme endonuclease III.Science. 1992 Oct 16;258(5081):434-40. doi: 10.1126/science.1411536. Science. 1992. PMID: 1411536
Cited by
-
Visualizing the Search for Radiation-damaged DNA Bases in Real Time.Radiat Phys Chem Oxf Engl 1993. 2016 Nov;128:126-133. doi: 10.1016/j.radphyschem.2016.05.011. Epub 2016 May 13. Radiat Phys Chem Oxf Engl 1993. 2016. PMID: 27818579 Free PMC article.
-
DNA damage recognition and repair by 3-methyladenine DNA glycosylase I (TAG).EMBO J. 2007 May 2;26(9):2411-20. doi: 10.1038/sj.emboj.7601649. Epub 2007 Apr 5. EMBO J. 2007. PMID: 17410210 Free PMC article.
-
Fluorescent probes for the analysis of DNA strand scission in base excision repair.Nucleic Acids Res. 2010 Apr;38(7):e101. doi: 10.1093/nar/gkq022. Epub 2010 Jan 27. Nucleic Acids Res. 2010. PMID: 20110254 Free PMC article.
-
Untwisting of the DNA helix stimulates the endonuclease activity of Bacillus subtilis Nth at AP sites.Nucleic Acids Res. 2012 Jan;40(2):739-50. doi: 10.1093/nar/gkr785. Epub 2011 Sep 27. Nucleic Acids Res. 2012. PMID: 21954439 Free PMC article.
-
Crystal structure and DNA cleavage mechanism of the restriction DNA glycosylase R.CcoLI from Campylobacter coli.Sci Rep. 2021 Jan 13;11(1):859. doi: 10.1038/s41598-020-79537-y. Sci Rep. 2021. PMID: 33441677 Free PMC article.
References
-
- Al-Tassan N. et al. (2002) Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat. Genet., 30, 227–232. - PubMed
-
- Asahara H., Wistort,P.M., Bank,J.F., Bakerian,R.H. and Cunningham,R.P. (1989) Purification and characterization of Escherichia coli endonuclease III from the cloned nth gene. Biochemistry, 28, 4444–4449. - PubMed
-
- Barrett T.E., Savva,R., Panayotou,G., Barlow,T., Brown,T., Jiricny,J. and Pearl,L.H. (1998) Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell, 92, 117–129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

