An active non-LTR retrotransposon with tandem structure in the compact genome of the pufferfish Tetraodon nigroviridis
- PMID: 12805276
- PMCID: PMC403742
- DOI: 10.1101/gr.726003
An active non-LTR retrotransposon with tandem structure in the compact genome of the pufferfish Tetraodon nigroviridis
Abstract
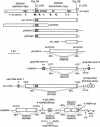
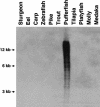
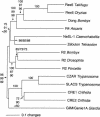
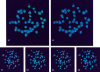
The fish retrotransposable element Zebulon encodes a reverse transcriptase and a carboxy-terminal restriction enzyme-like endonuclease, and is related phylogenetically to site-specific non-LTR retrotransposons from nematodes. Zebulon was detected in the pufferfishes Tetraodon nigroviridis and Takifugu rubripes, as well as in the zebrafish Danio rerio. Structural analysis suggested that Zebulon, in contrast to most non-LTR retrotransposons, might be able to retrotranspose as a partial tandem array. Zebulon was active relatively recently in the compact genome of T. nigroviridis, in which it contributed to the extension of intergenic and intronic sequences, and possibly to the formation of genomic rearrangements. Accumulation of Zebulon together with other retrotransposons was observed in some heterochromatic chromosomal regions of the genome of T. nigroviridis that might serve as reservoirs for active elements. Hence, pufferfish compact genomes are not evolutionarily inert and contain active retrotransposons, suggesting the presence of mechanisms allowing accumulation of retrotransposable elements in heterochromatin, but minimizing their impact on euchromatic regions. Homologous recombination between partial tandem sequences eliminating active copies of Zebulon and reducing the size of insertions in intronic and intragenic regions might represent such a mechanism.
Figures


Similar articles
-
Diversity and clustered distribution of retrotransposable elements in the compact genome of the pufferfish Tetraodon nigroviridis.Cytogenet Genome Res. 2005;110(1-4):522-36. doi: 10.1159/000084985. Cytogenet Genome Res. 2005. PMID: 16093705
-
Global heterochromatic colocalization of transposable elements with minisatellites in the compact genome of the pufferfish Tetraodon nigroviridis.Gene. 2004 Jul 21;336(2):175-83. doi: 10.1016/j.gene.2004.04.014. Gene. 2004. PMID: 15246529
-
Diversity of retrotransposable elements in compact pufferfish genomes.Trends Genet. 2003 Dec;19(12):674-8. doi: 10.1016/j.tig.2003.10.006. Trends Genet. 2003. PMID: 14642744 Review. No abstract available.
-
Non-LTR retrotransposons encoding a restriction enzyme-like endonuclease in vertebrates.J Mol Evol. 2001 Apr;52(4):351-60. doi: 10.1007/s002390010165. J Mol Evol. 2001. PMID: 11343131
-
DIRS-1 and the other tyrosine recombinase retrotransposons.Cytogenet Genome Res. 2005;110(1-4):575-88. doi: 10.1159/000084991. Cytogenet Genome Res. 2005. PMID: 16093711 Review.
Cited by
-
First insights on the retroelement Rex1 in the cytogenetics of frogs.Mol Cytogenet. 2015 Nov 5;8:86. doi: 10.1186/s13039-015-0189-5. eCollection 2015. Mol Cytogenet. 2015. PMID: 26550032 Free PMC article.
-
Chromosome spreading of associated transposable elements and ribosomal DNA in the fish Erythrinus erythrinus. Implications for genome change and karyoevolution in fish.BMC Evol Biol. 2010 Sep 6;10:271. doi: 10.1186/1471-2148-10-271. BMC Evol Biol. 2010. PMID: 20815941 Free PMC article.
-
Differential expression of a retrotransposable element, Rex6, in Colossoma macropomum fish from different Amazonian environments.Mob Genet Elements. 2014 Jul 21;4:e30003. doi: 10.4161/mge.30003. eCollection 2014. Mob Genet Elements. 2014. PMID: 25089227 Free PMC article.
-
Genomic organization of repetitive DNAs in the cichlid fish Astronotus ocellatus.Genetica. 2009 Jul;136(3):461-9. doi: 10.1007/s10709-008-9346-7. Epub 2008 Dec 27. Genetica. 2009. PMID: 19112556
-
Chromosome mapping of retrotransposable elements Rex1 and Rex3 in Leporinus Spix, 1829 species (Characiformes: Anostomidae) and its relationships among heterochromatic segments and W sex chromosome.Mob Genet Elements. 2013 Nov 1;3(6):e27460. doi: 10.4161/mge.27460. Epub 2013 Dec 10. Mob Genet Elements. 2013. PMID: 24404417 Free PMC article.
References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. - PubMed
-
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A.F., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301-1310. - PubMed
-
- Bartolomé, C., Maside, X., and Charlesworth, B. 2002. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol. Biol. Evol. 19: 926-937. - PubMed
-
- Boeke, J.D. and Chapman, K.B. 1991. Retrotransposition mechanisms. Curr. Opin. Cell Biol. 3: 502-507. - PubMed
WEB SITE REFERENCES
-
- http://fugu.hgmp.mrc.ac.uk/; The Fugu Genomics site at the UK HGMP Resource Centre.
-
- http://menu.hgmp.mrc.ac.uk/menu-bin/Nix; The Bio-informatics Application Server at the UK HGMP Resource Centre.
-
- http://www.ensembl.org/Danio_rerio/blastview; The Zebrafish BLAST server at the Wellcome Trust Sanger Institute.
-
- http://www.ncbi.nlm.nih.gov/BLAST; The BLAST server at the National Center for Biotechnology Information.
-
- http://www.ncbi.nlm.nih.gov/blast/tracemb.html; The Trace server at the National Center for Biotechnology information.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
