Dyskinetoplastic Trypanosoma brucei contains functional editing complexes
- PMID: 12796302
- PMCID: PMC161453
- DOI: 10.1128/EC.2.3.569-577.2003
Dyskinetoplastic Trypanosoma brucei contains functional editing complexes
Abstract
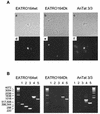
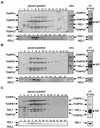
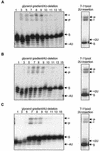
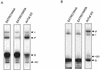
Mitochondrial pre-mRNAs undergo posttranscriptional RNA editing as directed by small guide RNAs (gRNAs) to produce functional mRNAs in kinetoplastid protozoa. The pre-mRNAs and gRNAs are encoded in the maxicircle and minicircle components, respectively, of the kinetoplastid mitochondrial DNA (kDNA), and editing is catalyzed by a multienzyme protein complex. Trypanosoma evansi AnTat3/3, which lacks maxicircles but retains a single class of minicircles, and a dyskinetoplastic mutant of Trypanosoma brucei EATRO164, which is devoid of kDNA, were both shown to retain genes and proteins for the editing complex. The proteins are present in complexes that immunoprecipitate and sediment indistinguishably from wild-type complexes. The complexes catalyze precleaved insertion and deletion editing as well as full-round deletion editing in vitro. Thus, mutants which lack the natural substrates for RNA editing and all or most gRNAs retain editing complexes that contain the four primary catalytic activities of editing and function in editing, at least in vitro. Therefore neither pre-mRNA nor gRNA is required to form functional RNA-editing complexes.
Figures

Similar articles
-
Processing of polycistronic guide RNAs is associated with RNA editing complexes in Trypanosoma brucei.EMBO J. 2000 Oct 16;19(20):5525-32. doi: 10.1093/emboj/19.20.5525. EMBO J. 2000. PMID: 11032819 Free PMC article.
-
Mitochondrial RNA editing in trypanosomes: small RNAs in control.Biochimie. 2014 May;100:125-31. doi: 10.1016/j.biochi.2014.01.003. Epub 2014 Jan 17. Biochimie. 2014. PMID: 24440637 Free PMC article. Review.
-
RNA editing in kinetoplastids.RNA Biol. 2010 Mar-Apr;7(2):229-36. doi: 10.4161/rna.7.2.11393. Epub 2010 Mar 1. RNA Biol. 2010. PMID: 20220308
-
Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria.EMBO J. 1997 Jul 1;16(13):4069-81. doi: 10.1093/emboj/16.13.4069. EMBO J. 1997. PMID: 9233816 Free PMC article.
-
Dual core processing: MRB1 is an emerging kinetoplast RNA editing complex.Trends Parasitol. 2013 Feb;29(2):91-9. doi: 10.1016/j.pt.2012.11.005. Epub 2013 Jan 8. Trends Parasitol. 2013. PMID: 23305619 Free PMC article. Review.
Cited by
-
KREH2 helicase represses ND7 mRNA editing in procyclic-stage Trypanosoma brucei by opposite modulation of canonical and 'moonlighting' gRNA utilization creating a proposed mRNA structure.Nucleic Acids Res. 2024 Oct 28;52(19):11940-11959. doi: 10.1093/nar/gkae699. Nucleic Acids Res. 2024. PMID: 39149912 Free PMC article.
-
The zinc-fingers of KREPA3 are essential for the complete editing of mitochondrial mRNAs in Trypanosoma brucei.PLoS One. 2010 Jan 27;5(1):e8913. doi: 10.1371/journal.pone.0008913. PLoS One. 2010. PMID: 20111718 Free PMC article.
-
Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates.Curr Genet. 2005 Nov;48(5):277-99. doi: 10.1007/s00294-005-0027-0. Epub 2005 Nov 4. Curr Genet. 2005. PMID: 16215758 Review.
-
Naphthalene-based RNA editing inhibitor blocks RNA editing activities and editosome assembly in Trypanosoma brucei.J Biol Chem. 2011 Apr 22;286(16):14178-89. doi: 10.1074/jbc.M110.199646. Epub 2011 Mar 4. J Biol Chem. 2011. PMID: 21378165 Free PMC article.
-
The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function.EMBO J. 2005 Dec 7;24(23):4029-40. doi: 10.1038/sj.emboj.7600862. Epub 2005 Nov 17. EMBO J. 2005. PMID: 16270030 Free PMC article.
References
-
- Aphasizhev, R., S. Sbicego, M. Peris, S. H. Jang, I. Aphasizheva, A. M. Simpson, A. Rivlin, and L. Simpson. 2002. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): the key enzyme in U-insertion/deletion RNA editing. Cell 108:637-648. - PubMed
-
- Bertrand, K. I., and S. L. Hajduk. 2000. Import of a constitutively expressed protein into mitochondria from procyclic and bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 106:249-260. - PubMed
-
- Blum, B., and L. Simpson. 1990. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell 62:391-397. - PubMed
-
- Borst, P., F. Fase-Fowler, and W. C. Gibson. 1987. Kinetoplast DNA of Trypanosoma evansi. Mol. Biochem. Parasitol. 23:31-38. - PubMed
-
- Borst, P., and J. H. J. Hoeijmakers. 1979. Extrachromosomal DNA. ICN-UCLA Symp. Mol. Cell. Biol. 15:533-548.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

