SPINE 2: a system for collaborative structural proteomics within a federated database framework
- PMID: 12771210
- PMCID: PMC156730
- DOI: 10.1093/nar/gkg397
SPINE 2: a system for collaborative structural proteomics within a federated database framework
Abstract
We present version 2 of the SPINE system for structural proteomics. SPINE is available over the web at http://nesg.org. It serves as the central hub for the Northeast Structural Genomics Consortium, allowing collaborative structural proteomics to be carried out in a distributed fashion. The core of SPINE is a laboratory information management system (LIMS) for key bits of information related to the progress of the consortium in cloning, expressing and purifying proteins and then solving their structures by NMR or X-ray crystallography. Originally, SPINE focused on tracking constructs, but, in its current form, it is able to track target sample tubes and store detailed sample histories. The core database comprises a set of standard relational tables and a data dictionary that form an initial ontology for proteomic properties and provide a framework for large-scale data mining. Moreover, SPINE sits at the center of a federation of interoperable information resources. These can be divided into (i) local resources closely coupled with SPINE that enable it to handle less standardized information (e.g. integrated mailing and publication lists), (ii) other information resources in the NESG consortium that are inter-linked with SPINE (e.g. crystallization LIMS local to particular laboratories) and (iii) international archival resources that SPINE links to and passes on information to (e.g. TargetDB at the PDB).
Figures
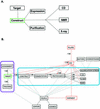
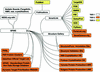



Similar articles
-
Laboratory information management system for membrane protein structure initiative--from gene to crystal.Mol Membr Biol. 2008 Dec;25(8):639-52. doi: 10.1080/09687680802511766. Mol Membr Biol. 2008. PMID: 18991141
-
SPINE: an integrated tracking database and data mining approach for identifying feasible targets in high-throughput structural proteomics.Nucleic Acids Res. 2001 Jul 1;29(13):2884-98. doi: 10.1093/nar/29.13.2884. Nucleic Acids Res. 2001. PMID: 11433035 Free PMC article.
-
LinkHub: a Semantic Web system that facilitates cross-database queries and information retrieval in proteomics.BMC Bioinformatics. 2007 May 9;8 Suppl 3(Suppl 3):S5. doi: 10.1186/1471-2105-8-S3-S5. BMC Bioinformatics. 2007. PMID: 17493288 Free PMC article. Review.
-
New York-Structural GenomiX Research Consortium (NYSGXRC): a large scale center for the protein structure initiative.J Struct Funct Genomics. 2005;6(2-3):225-32. doi: 10.1007/s10969-005-6827-0. J Struct Funct Genomics. 2005. PMID: 16211523
-
Data management in structural genomics: an overview.Methods Mol Biol. 2008;426:49-79. doi: 10.1007/978-1-60327-058-8_4. Methods Mol Biol. 2008. PMID: 18542857 Review.
Cited by
-
The high-throughput protein sample production platform of the Northeast Structural Genomics Consortium.J Struct Biol. 2010 Oct;172(1):21-33. doi: 10.1016/j.jsb.2010.07.011. Epub 2010 Aug 3. J Struct Biol. 2010. PMID: 20688167 Free PMC article.
-
A data management system for structural genomics.Proteome Sci. 2004 Jun 21;2(1):4. doi: 10.1186/1477-5956-2-4. Proteome Sci. 2004. PMID: 15210054 Free PMC article.
-
Predicting protein crystallization propensity from protein sequence.J Struct Funct Genomics. 2010 Mar;11(1):71-80. doi: 10.1007/s10969-010-9080-0. Epub 2010 Feb 23. J Struct Funct Genomics. 2010. PMID: 20177794 Free PMC article.
-
SpecDB: A relational database for archiving biomolecular NMR spectral data.J Magn Reson. 2022 Sep;342:107268. doi: 10.1016/j.jmr.2022.107268. Epub 2022 Jul 16. J Magn Reson. 2022. PMID: 35930941 Free PMC article.
-
Laboratory scale structural genomics.J Struct Funct Genomics. 2004;5(1-2):147-57. doi: 10.1023/B:JSFG.0000029193.82120.d1. J Struct Funct Genomics. 2004. PMID: 15263853
References
-
- Burley S.K. (2000) An overview of structural genomics. Nature Struct. Biol., 7 (Suppl.), 932–934. - PubMed
-
- Berman H.M., Bhat,T.N., Bourne,P.E., Feng,Z., Gilliland,G., Weissig,H. and Westbrook,J. (2000) The Protein Data Bank and the challenge of structural genomics. Nature Struct. Biol., 7 (Suppl.), 957–959. - PubMed
-
- Christendat D., Yee,A., Dharamsi,A., Kluger,Y., Savchenko,A., Cort,J.R., Booth,V., Mackereth,C.D., Saridakis,V., Ekiel,I. et al. (2000) Structural proteomics of an archaeon. Nature Struct. Biol., 7, 903–909. - PubMed
-
- Bertone P., Kluger,Y., Lan,N., Zheng,D., Christendat,D., Yee,A., Edwards,A.M., Arrowsmith,C.H., Montelione,G.T. and Gerstein,M. (2001) SPINE: an integrated tracking database and data mining approach for identifying feasible targets in high-throughput structural proteomics. Nucleic Acids Res., 29, 2884–2898. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

