CpG methylation of human papillomavirus type 16 DNA in cervical cancer cell lines and in clinical specimens: genomic hypomethylation correlates with carcinogenic progression
- PMID: 12743279
- PMCID: PMC154984
- DOI: 10.1128/jvi.77.11.6227-6234.2003
CpG methylation of human papillomavirus type 16 DNA in cervical cancer cell lines and in clinical specimens: genomic hypomethylation correlates with carcinogenic progression
Abstract
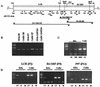
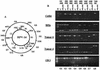
Infection with genital human papillomaviruses (HPVs) is the primary cause of cervical cancer. The infection is widespread, and little is known about the secondary factors associated with progression from subclinical infection to invasive carcinoma. Here we report that HPV genomes are efficiently targeted in vivo by CpG methylation, a well-known mechanism of transcriptional repression. Indeed, it has been shown previously that in vitro-methylated HPV type 16 (HPV-16) DNA is transcriptionally repressed after transfection into cell cultures. By using a scan with the restriction enzyme McrBC, we observed a conserved profile of CpG hyper- and hypomethylation throughout the HPV-16 genomes of the tumor-derived cell lines SiHa and CaSki. Methylation is particularly high in genomic segments overlying the late genes, while the long control region (LCR) and the oncogenes are unmethylated in the single HPV-16 copy in SiHa cells. In 81 patients from two different cohorts, the LCR and the E6 gene of HPV-16 DNA were found to be hypermethylated in 52% of asymptomatic smears, 21.7% of precursor lesions, and 6.1% of invasive carcinomas. This suggests that neoplastic transformation may be suppressed by CpG methylation, while demethylation occurs as the cause of or concomitant with neoplastic progression. These prevalences of hyper- and hypomethylation also indicate that CpG methylation plays an important role in the papillomavirus life cycle, which takes place in asymptomatic infections and precursor lesions but not in carcinomas. Bisulfite modification revealed that in most of the HPV-16 genomes of CaSki cells and of asymptomatic patients, all 11 CpG dinucleotides that overlap with the enhancer and the promoter were methylated, while in SiHa cells and cervical lesions, the same 11 or a subset of CpGs remained unmethylated. Our report introduces papillomaviruses as models to study the mechanism of CpG methylation, opens research on the importance of this mechanism during the viral life cycle, and provides a marker relevant for the etiology and diagnosis of cervical cancer.
Figures
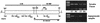
Similar articles
-
The human papillomavirus-18 genome is efficiently targeted by cellular DNA methylation.Virology. 2004 Jul 1;324(2):483-92. doi: 10.1016/j.virol.2004.04.002. Virology. 2004. PMID: 15207633
-
Methylation of the human papillomavirus-18 L1 gene: a biomarker of neoplastic progression?Virology. 2006 May 25;349(1):175-83. doi: 10.1016/j.virol.2005.12.033. Epub 2006 Feb 10. Virology. 2006. PMID: 16472835
-
CpG methylation in human papillomavirus (HPV) type 31 long control region (LCR) in cervical infections associated with cytological abnormalities.Virus Genes. 2016 Aug;52(4):552-5. doi: 10.1007/s11262-016-1338-6. Epub 2016 Apr 20. Virus Genes. 2016. PMID: 27098644
-
[Epidemiology of cervical papillomavirus infections. Recent knowledge].Presse Med. 2001 Jun 9;30(20):1017-23. Presse Med. 2001. PMID: 11433694 Review. French.
-
New markers for cervical dysplasia to visualise the genomic chaos created by aberrant oncogenic papillomavirus infections.Eur J Cancer. 2002 Nov;38(17):2229-42. doi: 10.1016/s0959-8049(02)00462-8. Eur J Cancer. 2002. PMID: 12441259 Review.
Cited by
-
The Hallmarks of Cervical Cancer: Molecular Mechanisms Induced by Human Papillomavirus.Biology (Basel). 2024 Jan 27;13(2):77. doi: 10.3390/biology13020077. Biology (Basel). 2024. PMID: 38392296 Free PMC article. Review.
-
HapMap methylation-associated SNPs, markers of germline DNA methylation, positively correlate with regional levels of human meiotic recombination.Genome Res. 2009 Apr;19(4):581-9. doi: 10.1101/gr.086181.108. Epub 2009 Jan 21. Genome Res. 2009. PMID: 19158364 Free PMC article.
-
Human papillomavirus 16 L1 gene methylation as a potential biomarker for predicting anal intraepithelial neoplasia in men who have sex with men (MSM).PLoS One. 2021 Sep 1;16(9):e0256852. doi: 10.1371/journal.pone.0256852. eCollection 2021. PLoS One. 2021. PMID: 34469465 Free PMC article.
-
Epigenetic Alterations in Human Papillomavirus-Associated Cancers.Viruses. 2017 Sep 1;9(9):248. doi: 10.3390/v9090248. Viruses. 2017. PMID: 28862667 Free PMC article. Review.
-
Activation of the retinoblastoma tumor suppressor mediates cell cycle inhibition and cell death in specific cervical cancer cell lines.Mol Carcinog. 2009 Jan;48(1):45-55. doi: 10.1002/mc.20456. Mol Carcinog. 2009. PMID: 18506774 Free PMC article.
References
-
- Baker, C. C., and C. Calef. 1995. Maps of papillomavirus transcripts, part III-A, p. 3-19. In G. Myers, H.-U. Bernard, H. Delius, C. Baker, J. Icenogle, A. Halpern, and C. Wheeler (ed.), Human papillomaviruses 1995 compendium. Los Alamos National Laboratory, Los Alamos, N.Mex.
-
- Bauer-Hofmann, R., C. Borghouts, E. Auvinen, E. Bourda, F. Rosl, and A. Alonso. 1996. Genomic cloning and characterization of the nonoccupied allele corresponding to the integration site of human papillomavirus type 16 DNA in the cervical cancer cell line SiHa. Virology 217:33-41. - PubMed
-
- Bernard, H.-U. 2000. Regulation of human papillomavirus gene expression by the factor CDP, nucleosomes and the nuclear matrix. Papillomavirus Rep. 11:73-80.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

