Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea
- PMID: 12732508
- PMCID: PMC154553
- DOI: 10.1128/AEM.69.5.2430-2443.2003
Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea
Abstract
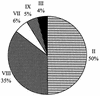
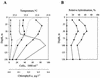
Phylogenetic relationships among members of the marine Synechococcus genus were determined following sequencing of the 16S ribosomal DNA (rDNA) from 31 novel cultured isolates from the Red Sea and several other oceanic environments. This revealed a large genetic diversity within the marine Synechococcus cluster consistent with earlier work but also identified three novel clades not previously recognized. Phylogenetic analyses showed one clade, containing halotolerant isolates lacking phycoerythrin (PE) and including strains capable, or not, of utilizing nitrate as the sole N source, which clustered within the MC-A (Synechococcus subcluster 5.1) lineage. Two copies of the 16S rRNA gene are present in marine Synechococcus genomes, and cloning and sequencing of these copies from Synechococcus sp. strain WH 7803 and genomic information from Synechococcus sp. strain WH 8102 reveal these to be identical. Based on the 16S rDNA sequence information, clade-specific oligonucleotides for the marine Synechococcus genus were designed and their specificity was optimized. Using dot blot hybridization technology, these probes were used to determine the in situ community structure of marine Synechococcus populations in the Red Sea at the time of a Synechococcus maximum during April 1999. A predominance of genotypes representative of a single clade was found, and these genotypes were common among strains isolated into culture. Conversely, strains lacking PE, which were also relatively easily isolated into culture, represented only a minor component of the Synechococcus population. Genotypes corresponding to well-studied laboratory strains also appeared to be poorly represented in this stratified water column in the Red Sea.
Figures





Similar articles
-
Closely related Prochlorococcus genotypes show remarkably different depth distributions in two oceanic regions as revealed by in situ hybridization using 16S rRNA-targeted oligonucleotides.Microbiology (Reading). 2001 Jul;147(Pt 7):1731-1744. doi: 10.1099/00221287-147-7-1731. Microbiology (Reading). 2001. PMID: 11429451
-
Colorful microdiversity of Synechococcus strains (picocyanobacteria) isolated from the Baltic Sea.ISME J. 2009 Apr;3(4):397-408. doi: 10.1038/ismej.2008.118. Epub 2008 Dec 4. ISME J. 2009. PMID: 19052629
-
Phylogenetic analyses of Synechococcus strains (cyanobacteria) using sequences of 16S rDNA and part of the phycocyanin operon reveal multiple evolutionary lines and reflect phycobilin content.Int J Syst Evol Microbiol. 2001 May;51(Pt 3):861-871. doi: 10.1099/00207713-51-3-861. Int J Syst Evol Microbiol. 2001. PMID: 11411708
-
Overview of the marine roseobacter lineage.Appl Environ Microbiol. 2005 Oct;71(10):5665-77. doi: 10.1128/AEM.71.10.5665-5677.2005. Appl Environ Microbiol. 2005. PMID: 16204474 Free PMC article. Review. No abstract available.
-
Picocyanobacterial Synechococcus in marine ecosystem: Insights from genetic diversity, global distribution, and potential function.Mar Environ Res. 2022 May;177:105622. doi: 10.1016/j.marenvres.2022.105622. Epub 2022 Apr 9. Mar Environ Res. 2022. PMID: 35429822 Review.
Cited by
-
Evolutionary Mechanisms of Long-Term Genome Diversification Associated With Niche Partitioning in Marine Picocyanobacteria.Front Microbiol. 2020 Sep 15;11:567431. doi: 10.3389/fmicb.2020.567431. eCollection 2020. Front Microbiol. 2020. PMID: 33042072 Free PMC article.
-
Phycoerythrin-specific bilin lyase-isomerase controls blue-green chromatic acclimation in marine Synechococcus.Proc Natl Acad Sci U S A. 2012 Dec 4;109(49):20136-41. doi: 10.1073/pnas.1211777109. Epub 2012 Nov 16. Proc Natl Acad Sci U S A. 2012. PMID: 23161909 Free PMC article.
-
Nested PCR Approach for petB Gene Metabarcoding of Marine Synechococcus Populations.Microbiol Spectr. 2023 Mar 6;11(2):e0408622. doi: 10.1128/spectrum.04086-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36877067 Free PMC article.
-
Diversity and evolution of phycobilisomes in marine Synechococcus spp.: a comparative genomics study.Genome Biol. 2007;8(12):R259. doi: 10.1186/gb-2007-8-12-r259. Genome Biol. 2007. PMID: 18062815 Free PMC article.
-
Seasonal dynamics in picocyanobacterial abundance and clade composition at coastal and offshore stations in the Baltic Sea.Sci Rep. 2022 Aug 22;12(1):14330. doi: 10.1038/s41598-022-18454-8. Sci Rep. 2022. PMID: 35995823 Free PMC article.
References
-
- Béjà, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. DeLong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2:516-529. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

