Distinct transcriptional pathways regulate basal and activated major histocompatibility complex class I expression
- PMID: 12724398
- PMCID: PMC154244
- DOI: 10.1128/MCB.23.10.3377-3391.2003
Distinct transcriptional pathways regulate basal and activated major histocompatibility complex class I expression
Abstract
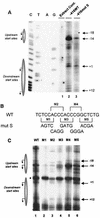
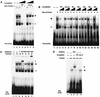
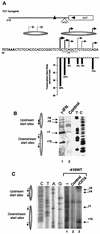
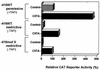
Transcription of major histocompatibility complex (MHC) class I genes is regulated by both tissue-specific (basal) and hormone/cytokine (activated) mechanisms. Although promoter-proximal regulatory elements have been characterized extensively, the role of the core promoter in mediating regulation has been largely undefined. We report here that the class I core promoter consists of distinct elements that are differentially utilized in basal and activated transcription pathways. These pathways recruit distinct transcription factor complexes to the core promoter elements and target distinct transcription initiation sites. Class I transcription initiates at four major sites within the core promoter and is clustered in two distinct regions: "upstream" (-14 and -18) and "downstream" (+12 and +1). Basal transcription initiates predominantly from the upstream start site region and is completely dependent upon the general transcription factor TAF1 (TAF(II)250). Activated transcription initiates predominantly from the downstream region and is TAF1 (TAF(II)250) independent. USF1 augments transcription initiating through the upstream start sites and is dependent on TAF1 (TAF(II)250), a finding consistent with its role in regulating basal class I transcription. In contrast, transcription activated by the interferon mediator CIITA is independent of TAF1 (TAF(II)250) and focuses initiation on the downstream start sites. Thus, basal and activated transcriptions of an MHC class I gene target distinct core promoter domains, nucleate distinct transcription initiation complexes and initiate at distinct sites within the promoter. We propose that transcription initiation at the core promoter is a dynamic process in which the mechanisms of core promoter function differ depending on the cellular environment.
Figures




Similar articles
-
Novel functions for TAF7, a regulator of TAF1-independent transcription.J Biol Chem. 2010 Dec 10;285(50):38772-80. doi: 10.1074/jbc.M110.173864. Epub 2010 Oct 11. J Biol Chem. 2010. PMID: 20937824 Free PMC article.
-
Three novel downstream promoter elements regulate MHC class I promoter activity in mammalian cells.PLoS One. 2010 Dec 13;5(12):e15278. doi: 10.1371/journal.pone.0015278. PLoS One. 2010. PMID: 21179443 Free PMC article.
-
TAF(II)250-independent transcription can be conferred on a TAF(II)250-dependent basal promoter by upstream activators.J Biol Chem. 2000 Apr 7;275(14):10160-7. doi: 10.1074/jbc.275.14.10160. J Biol Chem. 2000. PMID: 10744699
-
The general transcription machinery and general cofactors.Crit Rev Biochem Mol Biol. 2006 May-Jun;41(3):105-78. doi: 10.1080/10409230600648736. Crit Rev Biochem Mol Biol. 2006. PMID: 16858867 Review.
-
Regulation of gene expression via the core promoter and the basal transcriptional machinery.Dev Biol. 2010 Mar 15;339(2):225-9. doi: 10.1016/j.ydbio.2009.08.009. Epub 2009 Aug 13. Dev Biol. 2010. PMID: 19682982 Free PMC article. Review.
Cited by
-
Nonequivalence of classical MHC class I loci in ability to direct effective antiviral immunity.PLoS Pathog. 2012 Feb;8(2):e1002541. doi: 10.1371/journal.ppat.1002541. Epub 2012 Feb 23. PLoS Pathog. 2012. PMID: 22383876 Free PMC article.
-
Genomic characterization of MHC class I genes of the horse.Immunogenetics. 2005 Nov;57(10):763-74. doi: 10.1007/s00251-005-0034-9. Epub 2005 Nov 8. Immunogenetics. 2005. PMID: 16220348
-
Novel functions for TAF7, a regulator of TAF1-independent transcription.J Biol Chem. 2010 Dec 10;285(50):38772-80. doi: 10.1074/jbc.M110.173864. Epub 2010 Oct 11. J Biol Chem. 2010. PMID: 20937824 Free PMC article.
-
Tumor-induced escape mechanisms and their association with resistance to checkpoint inhibitor therapy.Cancer Immunol Immunother. 2019 Oct;68(10):1689-1700. doi: 10.1007/s00262-019-02373-1. Epub 2019 Aug 3. Cancer Immunol Immunother. 2019. PMID: 31375885 Free PMC article. Review.
-
A novel TBP-TAF complex on RNA polymerase II-transcribed snRNA genes.Transcription. 2012 Mar-Apr;3(2):92-104. doi: 10.4161/trns.19783. Epub 2012 Mar 1. Transcription. 2012. PMID: 22441827 Free PMC article.
References
-
- Aso, T., J. W. Conaway, and R. C. Conaway. 1994. Role of core promoter structure in assembly of the RNA polymerase II ormattedpreinitiation complex: a common pathway for formation of preinitiation intermediates at many TATA and TATA-less promoters. J. Biol. Chem. 269:26575-26583. - PubMed
-
- Berk, A. J. 1999. Activation of RNA polymerase II transcription. Curr. Opin. Cell Biol. 11:330-335. - PubMed
-
- Brodsky, F. M. 1999. Stealth, sabotage, and exploitation. Immunol. Rev. 168:5-11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
