Identification of essential residues involved in the allosteric modulation of the human A(3) adenosine receptor
- PMID: 12695530
- PMCID: PMC4367541
- DOI: 10.1124/mol.63.5.1021
Identification of essential residues involved in the allosteric modulation of the human A(3) adenosine receptor
Abstract
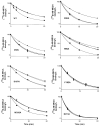
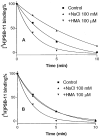
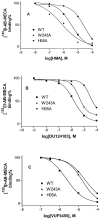
We examined the effects on allosteric modulation and ligand binding of the mutation of amino acid residues of the human A(3) adenosine receptor (A(3)AR) that are hypothesized to be near one of three loci: the putative sodium binding site, the putative ligand binding site, and the DRY motif in transmembrane helical domain 3. The effects of three heterocyclic allosteric modulators [the imidazoquinoline 2-cyclopentyl-4-phenylamino-1H-imidazo[4,5-c]quinoline (DU124183), the pyridinylisoquinoline 4-methoxy-N-[7-methyl-3-(2-pyridinyl)-1-isoquinolinyl]benzamide (VUF5455), and the amiloride analog 5-(N,N-hexamethylene)-amiloride] on the dissociation of the agonist radioligand, N(6)- (4-amino-3-[(125)I]iodobenzyl)-5'-N-methylcarboxamidoadenosine, were compared at wild-type (WT) and mutant A(3)ARs. The F182A(5.43) and N274A(7.45) mutations eliminated the allosteric effects of all three modulators but had little effect on agonist binding. The N30A(1.50) and D58N(2.50) mutations abolished the allosteric effects of DU124183 and VUF5455, but not HMA, whereas the D107N(3.49) mutation abolished the effects of DU124183, but not HMA or VUF5455. The T94A(3.36), H95A(3.37), K152A(EL2), W243A(6.48), L244A(6.49), and S247A(6.52) mutations did not influence allosteric effects of the modulators. Sodium ions (100 mM), which modulate agonist binding at a variety of receptors, caused an approximately 80% inhibition of agonist binding in WT A(3)ARs but did not show any effect on D58N(2.50), D107N(3.49), and F182A(5.43) mutant receptors. In contrast, NaCl induced a modest increase of agonist binding in N30A(1.50) and N274A(7.45) mutant receptors. NaCl decreased the dissociation rate of the antagonist radioligand [(3)H]8-ethyl-4-methyl-2-phenyl-(8R)-4,5,7,8-tetrahydro-1H-imidazo[2.1-i]purin-5-one (PSB-11) at the WT A(3)ARs, but not the D58N(2.50) mutant receptor. The results were interpreted using a rhodopsin-based molecular model of the A(3)AR to suggest multiple binding modes of the allosteric modulators.
Figures


Similar articles
-
Allosteric modulation of the adenosine family of receptors.Mini Rev Med Chem. 2005 Jun;5(6):545-53. doi: 10.2174/1389557054023242. Mini Rev Med Chem. 2005. PMID: 15974932 Free PMC article. Review.
-
Selective allosteric enhancement of agonist binding and function at human A3 adenosine receptors by a series of imidazoquinoline derivatives.Mol Pharmacol. 2002 Jul;62(1):81-9. doi: 10.1124/mol.62.1.81. Mol Pharmacol. 2002. PMID: 12065758 Free PMC article.
-
Derivatives of the triazoloquinazoline adenosine antagonist (CGS 15943) having high potency at the human A2B and A3 receptor subtypes.J Med Chem. 1998 Jul 16;41(15):2835-45. doi: 10.1021/jm980094b. J Med Chem. 1998. PMID: 9667972 Free PMC article.
-
Enabling Systemic Identification and Functionality Profiling for Cdc42 Homeostatic Modulators.bioRxiv [Preprint]. 2024 Jan 8:2024.01.05.574351. doi: 10.1101/2024.01.05.574351. bioRxiv. 2024. Update in: Commun Chem. 2024 Nov 19;7(1):271. doi: 10.1038/s42004-024-01352-7. PMID: 38260445 Free PMC article. Updated. Preprint.
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article. Review.
Cited by
-
Allosteric interactions at adenosine A(1) and A(3) receptors: new insights into the role of small molecules and receptor dimerization.Br J Pharmacol. 2014 Mar;171(5):1102-13. doi: 10.1111/bph.12345. Br J Pharmacol. 2014. PMID: 24024783 Free PMC article. Review.
-
Novobiocin and peptide analogs of α-factor are positive allosteric modulators of the yeast G protein-coupled receptor Ste2p.Biochim Biophys Acta. 2015 Apr;1848(4):916-24. doi: 10.1016/j.bbamem.2014.12.024. Epub 2015 Jan 7. Biochim Biophys Acta. 2015. PMID: 25576192 Free PMC article.
-
Structural basis for Na(+)-sensitivity in dopamine D2 and D3 receptors.Chem Commun (Camb). 2015 May 21;51(41):8618-21. doi: 10.1039/c5cc02204e. Chem Commun (Camb). 2015. PMID: 25896577 Free PMC article.
-
Keynote review: allosterism in membrane receptors.Drug Discov Today. 2006 Mar;11(5-6):191-202. doi: 10.1016/S1359-6446(05)03689-5. Drug Discov Today. 2006. PMID: 16580596 Free PMC article. Review.
-
Functional Characterization of Sodium Channel Inhibitors at the Delta-Opioid Receptor.ACS Omega. 2022 May 12;7(20):16939-16951. doi: 10.1021/acsomega.1c07226. eCollection 2022 May 24. ACS Omega. 2022. PMID: 35647460 Free PMC article.
References
-
- Angeli P, Marucci G, Buccioni M, Piergentili A, Giannella M, Quaglia W, Pigini M, Cantalamessa F, Nasuti C, Novi F, et al. Deoxamuscaroneoxime derivatives as useful muscarinic agonists to explore the muscarinic subsite: Demox, a modulator of orthosteric and allosteric sites at cardiac muscarinic M2 receptors. Life Sci. 2002;70:1427–1446. - PubMed
-
- Barbhaiya H, McClain R, IJzerman AP, Rivkees SA. Site-directed mutagenesis of the human A1 adenosine receptors: influences of acidic and hydroxy residues in the first four transmembrane domains on ligand binding. Mol Pharmacol. 1996;50:1635–1642. - PubMed
-
- Beukers MW, Kristiansen K, IJzerman AP, Edvardsen Ø. TinyGRAP database: a bioinformatics tool to mine G-protein-coupled receptor mutant data. Trends Pharmacol Sci. 1999;20:475–477. - PubMed
-
- Birdsall NJ, Lazareno S, Popham A, Saldanha J. Multiple allosteric sites on muscarinic receptors. Life Sci. 2001;68:2517–2524. - PubMed
-
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
