Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis
- PMID: 12692560
- PMCID: PMC1820765
- DOI: 10.1038/nbt816
Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis
Abstract
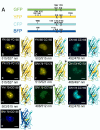
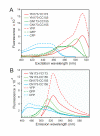
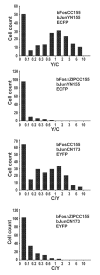
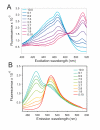
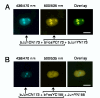
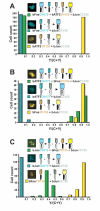
The specificity of biological regulatory mechanisms relies on selective interactions between different proteins in different cell types and in response to different extracellular signals. We describe a bimolecular fluorescence complementation (BiFC) approach for the simultaneous visualization of multiple protein interactions in the same cell. This approach is based on complementation between fragments of fluorescent proteins with different spectral characteristics. We have identified 12 bimolecular fluorescent complexes that correspond to 7 different spectral classes. Bimolecular complex formation between fragments of different fluorescent proteins did not differentially affect the dimerization efficiency of the bZIP domains of Fos and Jun or the subcellular sites of interactions between these domains. Multicolor BiFC enables visualization of interactions between different proteins in the same cell and comparison of the efficiencies of complex formation with alternative interaction partners.
Figures
Similar articles
-
Visualization of protein interactions in living cells using bimolecular fluorescence complementation (BiFC) analysis.Curr Protoc Cell Biol. 2006 Jan;Chapter 21:Unit 21.3. doi: 10.1002/0471143030.cb2103s29. Curr Protoc Cell Biol. 2006. PMID: 18228482
-
Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation.Mol Cell. 2002 Apr;9(4):789-98. doi: 10.1016/s1097-2765(02)00496-3. Mol Cell. 2002. PMID: 11983170
-
Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions.Biotechniques. 2006 Jan;40(1):61-6. doi: 10.2144/000112036. Biotechniques. 2006. PMID: 16454041
-
Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation.Chem Soc Rev. 2009 Oct;38(10):2876-86. doi: 10.1039/b909638h. Epub 2009 Sep 4. Chem Soc Rev. 2009. PMID: 19771334 Free PMC article. Review.
-
Transcription factors 1: bZIP proteins.Protein Profile. 1995;2(2):101-68. Protein Profile. 1995. PMID: 7780801 Review. No abstract available.
Cited by
-
Self-association of lymphocytic choriomeningitis virus nucleoprotein is mediated by its N-terminal region and is not required for its anti-interferon function.J Virol. 2012 Mar;86(6):3307-17. doi: 10.1128/JVI.05503-11. Epub 2012 Jan 18. J Virol. 2012. PMID: 22258244 Free PMC article.
-
In vivo BiFC analysis of Y14 and NXF1 mRNA export complexes: preferential localization within and around SC35 domains.J Cell Biol. 2006 Jan 30;172(3):373-81. doi: 10.1083/jcb.200503061. Epub 2006 Jan 23. J Cell Biol. 2006. PMID: 16431928 Free PMC article.
-
Development of bimolecular fluorescence complementation using Dronpa for visualization of protein-protein interactions in cells.Mol Imaging Biol. 2010 Oct;12(5):468-78. doi: 10.1007/s11307-010-0312-2. Mol Imaging Biol. 2010. PMID: 20373040
-
Bacterial effector binding to ribosomal protein s3 subverts NF-kappaB function.PLoS Pathog. 2009 Dec;5(12):e1000708. doi: 10.1371/journal.ppat.1000708. Epub 2009 Dec 24. PLoS Pathog. 2009. PMID: 20041225 Free PMC article.
-
A Live Cell Protein Complementation Assay for ORFeome-Wide Probing of Human HOX Interactomes.Cells. 2023 Jan 3;12(1):200. doi: 10.3390/cells12010200. Cells. 2023. PMID: 36611993 Free PMC article.
References
-
- Hu C-D, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. - PubMed
-
- Tsien RY. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. - PubMed
-
- Miyawaki A, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. - PubMed
-
- Romoser VA, Hinkle PM, Persechini A. Detection in living cells of Ca2+-dependent changes in the fluorescence emission of an indicator composed of two green fluorescent protein variants linked by a calmodulin-binding sequence. A new class of fluorescent indicators. J. Biol. Chem. 1997;272:13270–13274. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

