Endogenous assays of DNA methyltransferases: Evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo
- PMID: 12665573
- PMCID: PMC152572
- DOI: 10.1128/MCB.23.8.2709-2719.2003
Endogenous assays of DNA methyltransferases: Evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo
Abstract
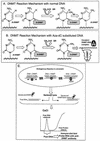
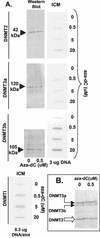
While CpG methylation can be readily analyzed at the DNA sequence level in wild-type and mutant cells, the actual DNA (cytosine-5) methyltransferases (DNMTs) responsible for in vivo methylation on genomic DNA are less tractable. We used an antibody-based method to identify specific endogenous DNMTs (DNMT1, DNMT1b, DNMT2, DNMT3a, and DNMT3b) that stably and selectively bind to genomic DNA containing 5-aza-2'-deoxycytidine (aza-dC) in vivo. Selective binding to aza-dC-containing DNA suggests that the engaged DNMT is catalytically active in the cell. DNMT1b is a splice variant of the predominant maintenance activity DNMT1, while DNMT2 is a well-conserved protein with homologs in plants, yeast, Drosophila, humans, and mice. Despite the presence of motifs essential for transmethylation activity, catalytic activity of DNMT2 has never been reported. The data here suggest that DNMT2 is active in vivo when the endogenous genome is the target, both in human and mouse cell lines. We quantified relative global genomic activity of DNMT1, -2, -3a, and -3b in a mouse teratocarcinoma cell line. DNMT1 and -3b displayed the greatest in vivo binding avidity for aza-dC-containing genomic DNA in these cells. This study demonstrates that individual DNMTs can be tracked and that their binding to genomic DNA can be quantified in mammalian cells in vivo. The different DNMTs display a wide spectrum of genomic DNA-directed activity. The use of an antibody-based tracking method will allow specific DNMTs and their DNA targets to be recovered and analyzed in a physiological setting in chromatin.
Figures




Similar articles
-
Comparative analysis of DNA methylation patterns in transgenic Drosophila overexpressing mouse DNA methyltransferases.Biochem J. 2004 Mar 15;378(Pt 3):763-8. doi: 10.1042/BJ20031567. Biochem J. 2004. PMID: 14636159 Free PMC article.
-
DNMT1, DNMT3A and DNMT3B proteins are differently expressed in mouse oocytes and early embryos.J Mol Histol. 2017 Dec;48(5-6):417-426. doi: 10.1007/s10735-017-9739-y. Epub 2017 Oct 13. J Mol Histol. 2017. PMID: 29027601
-
Targeting of 5-aza-2'-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme.Nucleic Acids Res. 2010 Jul;38(13):4313-24. doi: 10.1093/nar/gkq187. Epub 2010 Mar 25. Nucleic Acids Res. 2010. PMID: 20348135 Free PMC article.
-
[DNA methyltransferases: classification, functions and research progress].Yi Chuan. 2009 Sep;31(9):903-12. doi: 10.3724/sp.j.1005.2009.00903. Yi Chuan. 2009. PMID: 19819843 Review. Chinese.
-
[Dnmt2 is the Most Evolutionary Conserved and Enigmatic Cytosine DNA Methyltransferase in Eukaryotes].Genetika. 2016 Mar;52(3):269-82. Genetika. 2016. PMID: 27281847 Review. Russian.
Cited by
-
Norepinephrine, beyond the Synapse: Coordinating Epigenetic Codes for Memory.Int J Mol Sci. 2022 Aug 31;23(17):9916. doi: 10.3390/ijms23179916. Int J Mol Sci. 2022. PMID: 36077313 Free PMC article. Review.
-
Active DNA demethylation in post-mitotic neurons: a reason for optimism.Neuropharmacology. 2013 Dec;75:233-45. doi: 10.1016/j.neuropharm.2013.07.036. Epub 2013 Aug 16. Neuropharmacology. 2013. PMID: 23958448 Free PMC article. Review.
-
DNMT1 and DNMT3B regulate tumorigenicity of human prostate cancer cells by controlling RAD9 expression through targeted methylation.Carcinogenesis. 2021 Feb 25;42(2):220-231. doi: 10.1093/carcin/bgaa088. Carcinogenesis. 2021. PMID: 32780107 Free PMC article.
-
A clinical-molecular update on azanucleoside-based therapy for the treatment of hematologic cancers.Clin Epigenetics. 2016 Jun 21;8:71. doi: 10.1186/s13148-016-0237-y. eCollection 2016. Clin Epigenetics. 2016. PMID: 27330573 Free PMC article. Review.
-
Risk-association of DNA methyltransferases polymorphisms with gastric cancer in the Southern Chinese population.Int J Mol Sci. 2012;13(7):8364-8378. doi: 10.3390/ijms13078364. Epub 2012 Jul 5. Int J Mol Sci. 2012. PMID: 22942708 Free PMC article.
References
-
- Baylin, S. B., M. Esteller, M. R. Rountree, K. E. Bachman, K. Schuebel and J. G. Herman. 2001. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 10:687-692. - PubMed
-
- Bestor, T. 2000. The DNA methyltransferases of mammals. Hum. Mol. Genet. 9:2395-2402. - PubMed
-
- Bonfils, C., N. Beaulieu, E. Chan, J. Cotton-Montpetit, and A. R. MacLeod. 2000. Characterization of the human DNA methyltransferase splice variant Dnmt1b. J. Biol. Chem. 275:10754-10760. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
