Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator
- PMID: 12634378
- PMCID: PMC150665
- DOI: 10.1128/jvi.77.7.4205-4220.2003
Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator
Abstract
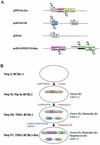
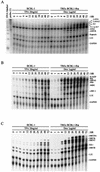
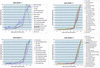
An important step in the herpesvirus life cycle is the switch from latency to lytic reactivation. In order to study the life cycle of Kaposi's sarcoma-associated herpesvirus (KSHV), we developed a gene expression system in KSHV-infected primary effusion lymphoma cells. This system uses Flp-mediated efficient recombination and tetracycline-inducible expression. The Rta transcriptional activator, which acts as a molecular switch for lytic reactivation of KSHV, was efficiently integrated downstream of the Flp recombination target site, and its expression was tightly controlled by tetracycline. Like stimulation with tetradecanoyl phorbol acetate (TPA), the ectopic expression of Rta efficiently induced a complete cycle of viral replication, including a well-ordered program of KSHV gene expression and production of infectious viral progeny. A striking feature of Rta-mediated lytic gene expression was that Rta induced KSHV gene expression in a more powerful and efficient manner than TPA stimulation, indicating that Rta plays a central, leading role in KSHV lytic gene expression. Thus, our streamlined gene expression system provides a novel means not only to study the effects of viral gene products on overall KSHV gene expression and replication, but also to understand the natural viral reactivation process.
Figures
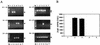



Similar articles
-
CCAAT/enhancer-binding protein-alpha is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters.J Virol. 2003 Sep;77(17):9590-612. doi: 10.1128/jvi.77.17.9590-9612.2003. J Virol. 2003. PMID: 12915572 Free PMC article.
-
ARID3B: a Novel Regulator of the Kaposi's Sarcoma-Associated Herpesvirus Lytic Cycle.J Virol. 2016 Sep 29;90(20):9543-55. doi: 10.1128/JVI.03262-15. Print 2016 Oct 15. J Virol. 2016. PMID: 27512077 Free PMC article.
-
Comparative study of regulation of RTA-responsive genes in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8.J Virol. 2003 Sep;77(17):9451-62. doi: 10.1128/jvi.77.17.9451-9462.2003. J Virol. 2003. PMID: 12915560 Free PMC article.
-
The Rta/Orf50 transactivator proteins of the gamma-herpesviridae.Curr Top Microbiol Immunol. 2007;312:71-100. doi: 10.1007/978-3-540-34344-8_3. Curr Top Microbiol Immunol. 2007. PMID: 17089794 Review.
-
Lytic cycle switches of oncogenic human gammaherpesviruses.Adv Cancer Res. 2007;97:81-109. doi: 10.1016/S0065-230X(06)97004-3. Adv Cancer Res. 2007. PMID: 17419942 Review.
Cited by
-
MicroRNAs and unusual small RNAs discovered in Kaposi's sarcoma-associated herpesvirus virions.J Virol. 2012 Dec;86(23):12717-30. doi: 10.1128/JVI.01473-12. Epub 2012 Sep 12. J Virol. 2012. PMID: 22973026 Free PMC article.
-
Quantitative analysis of the bidirectional viral G-protein-coupled receptor and lytic latency-associated nuclear antigen promoter of Kaposi's sarcoma-associated herpesvirus.J Virol. 2012 Sep;86(18):9683-95. doi: 10.1128/JVI.00881-12. Epub 2012 Jun 27. J Virol. 2012. PMID: 22740392 Free PMC article.
-
Fine-Tuning of the Kaposi's Sarcoma-Associated Herpesvirus Life Cycle in Neighboring Cells through the RTA-JAG1-Notch Pathway.PLoS Pathog. 2016 Oct 19;12(10):e1005900. doi: 10.1371/journal.ppat.1005900. eCollection 2016 Oct. PLoS Pathog. 2016. PMID: 27760204 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus K7 protein targets a ubiquitin-like/ubiquitin-associated domain-containing protein to promote protein degradation.Mol Cell Biol. 2004 May;24(9):3938-48. doi: 10.1128/MCB.24.9.3938-3948.2004. Mol Cell Biol. 2004. PMID: 15082787 Free PMC article.
-
Non-human primate model of Kaposi's sarcoma-associated herpesvirus infection.PLoS Pathog. 2009 Oct;5(10):e1000606. doi: 10.1371/journal.ppat.1000606. Epub 2009 Oct 2. PLoS Pathog. 2009. PMID: 19798430 Free PMC article.
References
-
- Alexander, L., L. Denekamp, A. Knapp, M. R. Auerbach, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388-3398. - PMC - PubMed
-
- Ambroziak, J. A., D. J. Blackbourn, B. G. Herndier, R. G. Glogau, J. H. Gullett, A. R. McDonald, E. T. Lennette, and J. A. Levy. 1995. Herpes-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science 268:582-583. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

