Distinct mechanisms of neutralization by monoclonal antibodies specific for sites in the N-terminal or C-terminal domain of murine leukemia virus SU
- PMID: 12634359
- PMCID: PMC150638
- DOI: 10.1128/jvi.77.7.3993-4003.2003
Distinct mechanisms of neutralization by monoclonal antibodies specific for sites in the N-terminal or C-terminal domain of murine leukemia virus SU
Abstract
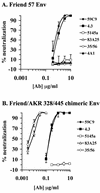
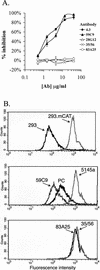
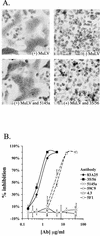
The epitope specificities and functional activities of monoclonal antibodies (MAbs) specific for the murine leukemia virus (MuLV) SU envelope protein subunit were determined. Neutralizing antibodies were directed towards two distinct sites in MuLV SU: one overlapping the major receptor-binding pocket in the N-terminal domain and the other involving a region that includes the most C-terminal disulfide-bonded loop. Two other groups of MAbs, reactive with distinct sites in the N-terminal domain or in the proline-rich region (PRR), did not neutralize MuLV infectivity. Only the neutralizing MAbs specific for the receptor-binding pocket were able to block binding of purified SU and MuLV virions to cells expressing the ecotropic MuLV receptor, mCAT-1. Whereas the neutralizing MAbs specific for the C-terminal domain did not interfere with the SU-mCAT-1 interaction, they efficiently inhibited cell-to-cell fusion mediated by MuLV Env, indicating that they interfered with a postattachment event necessary for fusion. The C-terminal domain MAbs displayed the highest neutralization titers and binding activities. However, the nonneutralizing PRR-specific MAbs bound to intact virions with affinities similar to those of the neutralizing receptor-binding pocket-specific MAbs, indicating that epitope exposure, while necessary, is not sufficient for viral neutralization by MAbs. These results identify two separate neutralization domains in MuLV SU and suggest a role for the C-terminal domain in a postattachment step necessary for viral fusion.
Figures



Similar articles
-
Involvement of the C-terminal disulfide-bonded loop of murine leukemia virus SU protein in a postbinding step critical for viral entry.J Virol. 2005 Jun;79(12):7868-76. doi: 10.1128/JVI.79.12.7868-7876.2005. J Virol. 2005. PMID: 15919941 Free PMC article.
-
Properties of the naturally occurring soluble surface glycoprotein of ecotropic murine leukemia virus: binding specificity and possible conformational change after binding to receptor.J Virol. 2000 Feb;74(4):1815-26. doi: 10.1128/jvi.74.4.1815-1826.2000. J Virol. 2000. PMID: 10644355 Free PMC article.
-
Effects of subtle changes in the SU protein of ecotropic murine leukemia virus on its brain capillary endothelial cell tropism and interference properties.Virology. 1996 Jan 15;215(2):142-51. doi: 10.1006/viro.1996.0017. Virology. 1996. PMID: 8560761
-
The glycoprotein G of rhabdoviruses.Arch Virol. 1995;140(5):827-51. doi: 10.1007/BF01314961. Arch Virol. 1995. PMID: 7605197 Review.
-
Occupancy and mechanism in antibody-mediated neutralization of animal viruses.J Gen Virol. 2002 Sep;83(Pt 9):2091-2108. doi: 10.1099/0022-1317-83-9-2091. J Gen Virol. 2002. PMID: 12185262 Review.
Cited by
-
Involvement of the C-terminal disulfide-bonded loop of murine leukemia virus SU protein in a postbinding step critical for viral entry.J Virol. 2005 Jun;79(12):7868-76. doi: 10.1128/JVI.79.12.7868-7876.2005. J Virol. 2005. PMID: 15919941 Free PMC article.
-
Antibody-induced conformational changes in herpes simplex virus glycoprotein gD reveal new targets for virus neutralization.J Virol. 2012 Feb;86(3):1563-76. doi: 10.1128/JVI.06480-11. Epub 2011 Nov 30. J Virol. 2012. PMID: 22130533 Free PMC article.
-
The fusion-controlling disulfide bond isomerase in retrovirus Env is triggered by protein destabilization.J Virol. 2005 Feb;79(3):1678-85. doi: 10.1128/JVI.79.3.1678-1685.2005. J Virol. 2005. PMID: 15650193 Free PMC article.
-
Neutralization of zoonotic retroviruses by human antibodies: Genotype-specific epitopes within the receptor-binding domain from simian foamy virus.PLoS Pathog. 2023 Apr 24;19(4):e1011339. doi: 10.1371/journal.ppat.1011339. eCollection 2023 Apr. PLoS Pathog. 2023. PMID: 37093892 Free PMC article.
-
Unique Structure and Distinctive Properties of the Ancient and Ubiquitous Gamma-Type Envelope Glycoprotein.Viruses. 2023 Jan 18;15(2):274. doi: 10.3390/v15020274. Viruses. 2023. PMID: 36851488 Free PMC article. Review.
References
-
- Albritton, L. M., L. Tseng, D. Scadden, and J. M. Cunningham. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659-666. - PubMed
-
- Burkhart, M. D. 2002. A study of retroviral neutralization by monoclonal antibodies specific for the murine leukemia virus envelope subunit SU. Ph.D. dissertation. New York University, New York, N.Y.
-
- Burton, D., E. Saphire, and P. Parren. 2001. A model for neutralization of viruses based on antibody coating of the virion surface, p. 109-143. In D. Burton (ed.), Antibodies in viral infection. Springer-Verlag, Berlin, Germany. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

