Genetic diversity: frameshift mechanisms alter coding of a gene (Epstein-Barr virus LF3 gene) that contains multiple 102-base-pair direct sequence repeats
- PMID: 12612089
- PMCID: PMC149476
- DOI: 10.1128/MCB.23.6.2192-2201.2003
Genetic diversity: frameshift mechanisms alter coding of a gene (Epstein-Barr virus LF3 gene) that contains multiple 102-base-pair direct sequence repeats
Abstract
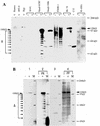
Frameshift mutations provide recognized mechanisms for changing the coding potential of an organism. Here, multiple frameshifts are identified in repetitive sequences within an Epstein-Barr virus unspliced early gene, LF3, which is associated with the viral replicative cycle and also transcriptionally expressed in many virally associated tumors. On the DNA strand encoding LF3, there are three open reading frames, only one of which contains an initiation codon. Most (>95%) of the gene consists of numerous (>20, varying with cell source) GC-rich copies of a 102-bp direct repeat (called IR 4) flanked by small unique sequences. LF3 may express a protein if its initiation and termination codons reside in the same reading frame, but this is not always the case. Frameshifting events, occurring in short runs of pyrimidines (mainly C residues) in the repeats, give rise to mutations which may provide a mechanism for escape of an LF3 function from host surveillance. Sequence studies link these frameshifts to DNA replication errors. Notably, the number of sites in LF3 at which such mutations can occur permits a very large amount of diversity in this gene. Our data also suggest a second degeneracy mechanism within the protein itself, which influences its stability and may reflect a host defense mechanism. LF3 thus provides a potentially important model for studying the quest for supremacy between a virus and its host.
Figures



Similar articles
-
Expression of two related viral early genes in Epstein-Barr virus-associated tumors.J Virol. 2000 Mar;74(6):2793-803. doi: 10.1128/jvi.74.6.2793-2803.2000. J Virol. 2000. PMID: 10684296 Free PMC article.
-
Epstein-Barr virus latent membrane protein-1 oncogene deletions: correlations with malignancy in Epstein-Barr virus--associated lymphoproliferative disorders and malignant lymphomas.Blood. 1996 Jul 1;88(1):242-51. Blood. 1996. PMID: 8704180
-
A basis for new approaches to the chemotherapy of AIDS: novel genes in HIV-1 potentially encode selenoproteins expressed by ribosomal frameshifting and termination suppression.J Med Chem. 1994 Aug 19;37(17):2637-54. doi: 10.1021/jm00043a004. J Med Chem. 1994. PMID: 8064794
-
[Epstein-Barr virus].Nihon Rinsho. 2003 Mar;61 Suppl 3:582-6. Nihon Rinsho. 2003. PMID: 12718032 Review. Japanese. No abstract available.
-
[Structural organization of the Epstein-Barr virus genome].Eksp Onkol. 1984;6(4):3-10. Eksp Onkol. 1984. PMID: 6209094 Review. Russian.
Cited by
-
Long non-coding RNAs in Epstein-Barr virus-related cancer.Cancer Cell Int. 2021 May 25;21(1):278. doi: 10.1186/s12935-021-01986-w. Cancer Cell Int. 2021. PMID: 34034760 Free PMC article. Review.
-
The Spike Protein of SARS-coV2 19B (S) Clade Mirrors Critical Features of Viral Adaptation and Coevolution.Microorganisms. 2022 Oct 12;10(10):2017. doi: 10.3390/microorganisms10102017. Microorganisms. 2022. PMID: 36296293 Free PMC article.
-
Early Epstein-Barr Virus Genomic Diversity and Convergence toward the B95.8 Genome in Primary Infection.J Virol. 2018 Jan 2;92(2):e01466-17. doi: 10.1128/JVI.01466-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29093087 Free PMC article.
-
Complexities associated with expression of Epstein-Barr virus (EBV) lytic origins of DNA replication.Nucleic Acids Res. 2007;35(10):3391-406. doi: 10.1093/nar/gkm170. Epub 2007 May 3. Nucleic Acids Res. 2007. PMID: 17478522 Free PMC article.
-
Regulators of Viral Frameshifting: More Than RNA Influences Translation Events.Annu Rev Virol. 2020 Sep 29;7(1):219-238. doi: 10.1146/annurev-virology-012120-101548. Epub 2020 Jun 29. Annu Rev Virol. 2020. PMID: 32600156 Free PMC article. Review.
References
-
- Boyer, J. C., K. Bebenedk, and T. A. Kunkel. 1996. Analyzing the fidelity of reverse transcription and translation. Methods Enzymol. 275:523-537. - PubMed
-
- Busson, P., G. Ganem, P. Flores, F. Mugneret, B. Clausse, B. Caillou, K. Brahan, et al. 1988. Establishment and characterization of three transplantable EBV-containing nasopharyngeal carcinoma. Int. J. Cancer 42:599-606. - PubMed
-
- Cattaneo, R., K. Kaelin, K. Baczko, and M. Billeter. 1989. Measles virus editing provides an additional cysteine-rich protein. Cell 56:759-764. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
