Activation of the early B-cell-specific mb-1 (Ig-alpha) gene by Pax-5 is dependent on an unmethylated Ets binding site
- PMID: 12612069
- PMCID: PMC149480
- DOI: 10.1128/MCB.23.6.1946-1960.2003
Activation of the early B-cell-specific mb-1 (Ig-alpha) gene by Pax-5 is dependent on an unmethylated Ets binding site
Abstract
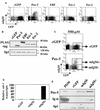
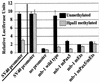
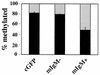
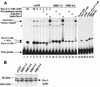
Methylation of cytosine in CpG dinucleotides promotes transcriptional repression in mammals by blocking transcription factor binding and recruiting methyl-binding proteins that initiate chromatin remodeling. Here, we use a novel cell-based system to show that retrovirally expressed Pax-5 protein activates endogenous early B-cell-specific mb-1 genes in plasmacytoma cells, but only when the promoter is hypomethylated. CpG methylation does not directly affect binding of the promoter by Pax-5. Instead, methylation of an adjacent CpG interferes with assembly of ternary complexes comprising Pax-5 and Ets proteins. In electrophoretic mobility shift assays, recruitment of Ets-1 is blocked by methylation of the Ets site (5'CCGGAG) on the antisense strand. In transfection assays, selective methylation of a single CpG within the Pax-5-dependent Ets site greatly reduces mb-1 promoter activity. Prior demethylation of the endogenous mb-1 promoter is required for its activation by Pax-5 in transduced cells. Although B-lineage cells have only unmethylated mb-1 genes and do not modulate methylation of the mb-1 promoter during development, other tissues feature high percentages of methylated alleles. Together, these studies demonstrate a novel DNA methylation-dependent mechanism for regulating transcriptional activity through the inhibition of DNA-dependent protein-protein interactions.
Figures



Similar articles
-
Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter.Genes Dev. 1996 Sep 1;10(17):2198-211. doi: 10.1101/gad.10.17.2198. Genes Dev. 1996. PMID: 8804314
-
Requirements for selective recruitment of Ets proteins and activation of mb-1/Ig-alpha gene transcription by Pax-5 (BSAP).Nucleic Acids Res. 2003 Oct 1;31(19):5483-9. doi: 10.1093/nar/gkg785. Nucleic Acids Res. 2003. PMID: 14500810 Free PMC article.
-
Highly conserved amino acids in Pax and Ets proteins are required for DNA binding and ternary complex assembly.Nucleic Acids Res. 2001 Oct 15;29(20):4154-65. doi: 10.1093/nar/29.20.4154. Nucleic Acids Res. 2001. PMID: 11600704 Free PMC article.
-
Regulation of Ets function by protein - protein interactions.Oncogene. 2000 Dec 18;19(55):6514-23. doi: 10.1038/sj.onc.1204035. Oncogene. 2000. PMID: 11175367 Review.
-
Ets-1 flips for new partner Pax-5.Structure. 2002 Jan;10(1):11-4. doi: 10.1016/s0969-2126(01)00701-8. Structure. 2002. PMID: 11796106 Review.
Cited by
-
Association of E26 Transformation Specific Sequence 1 Variants with Rheumatoid Arthritis in Chinese Han Population.PLoS One. 2015 Aug 4;10(8):e0134875. doi: 10.1371/journal.pone.0134875. eCollection 2015. PLoS One. 2015. PMID: 26241881 Free PMC article.
-
Early B-Cell Factor 1: An Archetype for a Lineage-Restricted Transcription Factor Linking Development to Disease.Adv Exp Med Biol. 2024;1459:143-156. doi: 10.1007/978-3-031-62731-6_7. Adv Exp Med Biol. 2024. PMID: 39017843 Review.
-
Ikaros promotes rearrangement of TCR α genes in an Ikaros null thymoma cell line.Eur J Immunol. 2013 Feb;43(2):521-32. doi: 10.1002/eji.201242757. Epub 2012 Dec 27. Eur J Immunol. 2013. PMID: 23172374 Free PMC article.
-
Regulation of cancer germline antigen gene expression: implications for cancer immunotherapy.Future Oncol. 2010 May;6(5):717-32. doi: 10.2217/fon.10.36. Future Oncol. 2010. PMID: 20465387 Free PMC article. Review.
-
Tissue-Specific Expression of Estrogen Receptor 1 Is Regulated by DNA Methylation in a T-DMR.Mol Endocrinol. 2016 Mar;30(3):335-47. doi: 10.1210/me.2015-1058. Epub 2015 Dec 18. Mol Endocrinol. 2016. PMID: 26683811 Free PMC article.
References
-
- Adams, B., P. Dorfler, A. Aguzzi, Z. Kozmik, P. Urbanek, I. Maurer-Fogy, and M. Busslinger. 1992. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 6:1589-1607. - PubMed
-
- Bird, A. P., and A. P. Wolffe. 1999. Methylation-induced repression—belts, braces and chromatin. Cell 99:451-454. - PubMed
-
- Burger, C., and A. Radbruch. 1990. Protective methylation of immunoglobulin and T cell receptor (TcR) gene loci prior to induction of class switch and TcR recombination. Eur. J. Immunol. 20:2285-2291. - PubMed
-
- Burgers, W. A., F. Fuks, and T. Kouzarides. 2002. DNA methyltransferases get connected to chromatin. Trends Genet. 18:275-277. - PubMed
-
- Busslinger, M., and P. Urbanek. 1995. The role of BSAP (Pax-5) in B-cell development. Curr. Opin. Genet. Dev. 5:595-601. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
