Genetic and biochemical analysis of phosphatase activity of Escherichia coli NRII (NtrB) and its regulation by the PII signal transduction protein
- PMID: 12562801
- PMCID: PMC142841
- DOI: 10.1128/JB.185.4.1299-1315.2003
Genetic and biochemical analysis of phosphatase activity of Escherichia coli NRII (NtrB) and its regulation by the PII signal transduction protein
Abstract
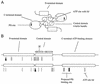
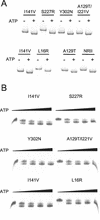
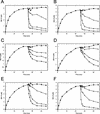
Mutant forms of Escherichia coli NRII (NtrB) were isolated that retained wild-type NRII kinase activity but were defective in the PII-activated phosphatase activity of NRII. Mutant strains were selected as mimicking the phenotype of a strain (strain BK) that lacks both of the related PII and GlnK signal transduction proteins and thus has no mechanism for activation of the NRII phosphatase activity. The selection and screening procedure resulted in the isolation of numerous mutants that phenotypically resembled strain BK to various extents. Mutations mapped to the glnL (ntrB) gene encoding NRII and were obtained in all three domains of NRII. Two distinct regions of the C-terminal, ATP-binding domain were identified by clusters of mutations. One cluster, including the Y302N mutation, altered a lid that sits over the ATP-binding site of NRII. The other cluster, including the S227R mutation, defined a small surface on the "back" or opposite side of this domain. The S227R and Y302N proteins were purified, along with the A129T (NRII2302) protein, which has reduced phosphatase activity due to a mutation in the central domain of NRII, and the L16R protein, which has a mutation in the N-terminal domain of NRII. The S227R, Y302N, and L16R proteins were specifically defective in the PII-activated phosphatase activity of NRII. Wild-type NRII, Y302N, A129T, and L16R proteins bound to PII, while the S227R protein was defective in binding PII. This suggests that the PII-binding site maps to the "back" of the C-terminal domain and that mutation of the ATP-lid, central domain, and N-terminal domain altered functions necessary for the phosphatase activity after PII binding.
Figures



Similar articles
-
Mechanism of the PII-activated phosphatase activity of Escherichia coli NRII (NtrB): how the different domains of NRII collaborate to act as a phosphatase.Biochemistry. 2003 Jul 29;42(29):8885-99. doi: 10.1021/bi030065p. Biochemistry. 2003. PMID: 12873150
-
Functional dissection of the dimerization and enzymatic activities of Escherichia coli nitrogen regulator II and their regulation by the PII protein.Biochemistry. 2000 Nov 7;39(44):13433-49. doi: 10.1021/bi000794u. Biochemistry. 2000. PMID: 11063580
-
Crystal structure of the C-terminal domain of the two-component system transmitter protein nitrogen regulator II (NRII; NtrB), regulator of nitrogen assimilation in Escherichia coli.Biochemistry. 2004 Jun 1;43(21):6670-8. doi: 10.1021/bi049474r. Biochemistry. 2004. PMID: 15157101
-
Mechanism of regulation of the bifunctional histidine kinase NtrB in Escherichia coli.J Mol Microbiol Biotechnol. 2002 May;4(3):229-33. J Mol Microbiol Biotechnol. 2002. PMID: 11931552 Review.
-
Integration of antagonistic signals in the regulation of nitrogen assimilation in Escherichia coli.Curr Top Cell Regul. 2000;36:31-75. doi: 10.1016/s0070-2137(01)80002-9. Curr Top Cell Regul. 2000. PMID: 10842746 Review. No abstract available.
Cited by
-
Bacterial Histidine Kinase and the Development of Its Inhibitors in the 21st Century.Antibiotics (Basel). 2024 Jun 22;13(7):576. doi: 10.3390/antibiotics13070576. Antibiotics (Basel). 2024. PMID: 39061258 Free PMC article. Review.
-
Mutations altering the N-terminal receiver domain of NRI (NtrC) That prevent dephosphorylation by the NRII-PII complex in Escherichia coli.J Bacteriol. 2004 Sep;186(17):5730-40. doi: 10.1128/JB.186.17.5730-5740.2004. J Bacteriol. 2004. PMID: 15317778 Free PMC article.
-
Negative control in two-component signal transduction by transmitter phosphatase activity.Mol Microbiol. 2011 Oct;82(2):275-86. doi: 10.1111/j.1365-2958.2011.07829.x. Epub 2011 Sep 29. Mol Microbiol. 2011. PMID: 21895797 Free PMC article. Review.
-
Specificity residues determine binding affinity for two-component signal transduction systems.mBio. 2013 Nov 5;4(6):e00420-13. doi: 10.1128/mBio.00420-13. mBio. 2013. PMID: 24194534 Free PMC article.
-
How important is the phosphatase activity of sensor kinases?Curr Opin Microbiol. 2010 Apr;13(2):168-76. doi: 10.1016/j.mib.2010.01.013. Epub 2010 Mar 10. Curr Opin Microbiol. 2010. PMID: 20223700 Free PMC article. Review.
References
-
- Atkinson, M. R., E. S. Kamberov, R. L. Weiss, and A. J. Ninfa. 1994. Reversible uridylylation of the Escherichia coli PII signal transduction protein regulates its ability to stimulate the dephosphorylation of the transcription factor nitrogen regulator I (NRI or NtrC). J. Biol. Chem. 269:28288-28293. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

