Yeast DNA polymerase zeta is an efficient extender of primer ends opposite from 7,8-dihydro-8-Oxoguanine and O6-methylguanine
- PMID: 12556503
- PMCID: PMC141155
- DOI: 10.1128/MCB.23.4.1453-1459.2003
Yeast DNA polymerase zeta is an efficient extender of primer ends opposite from 7,8-dihydro-8-Oxoguanine and O6-methylguanine
Abstract
Genetic studies in Saccharomyces cerevisiae have indicated the requirement of DNA polymerase (Pol) zeta for mutagenesis induced by UV light and by other DNA damaging agents. However, on its own, Pol zeta is highly inefficient at replicating through DNA lesions; rather, it promotes their mutagenic bypass by extending from the nucleotide inserted opposite the lesion by another DNA polymerase. So far, such a role for Pol zeta has been established for cyclobutane pyrimidine dimers, (6-4) dipyrimidine photoproducts, and abasic sites. Here, we examine whether Pol zeta can replicate through the 7,8-dihydro-8-oxoguanine (8-oxoG) and O(6)-methylguanine (m6G) lesions. We chose these two lesions for this study because the replicative polymerase, Pol delta, can replicate through them, albeit weakly. We found that Pol zeta is very inefficient at inserting nucleotides opposite both these lesions, but it can efficiently extend from the nucleotides inserted opposite them by Pol delta. Also, the most efficient bypass of 8-oxoG and m6G lesions occurs when Pol delta is combined with Pol zeta, indicating a role for Polzeta in extending from the nucleotides inserted opposite these lesions by Pol delta. Thus, Pol zeta is a highly specialized polymerase that can proficiently extend from the primer ends opposite DNA lesions, irrespective of their degree of geometric distortion. Pol zeta, however, is unusually sensitive to geometric distortion of the templating residue, as it is highly inefficient at incorporating nucleotides even opposite the moderately distorting 8-oxoG and m6G lesions.
Figures
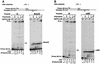

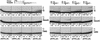
Similar articles
-
Mechanism of efficient and accurate nucleotide incorporation opposite 7,8-dihydro-8-oxoguanine by Saccharomyces cerevisiae DNA polymerase eta.Mol Cell Biol. 2005 Mar;25(6):2169-76. doi: 10.1128/MCB.25.6.2169-2176.2005. Mol Cell Biol. 2005. PMID: 15743815 Free PMC article.
-
Role of human DNA polymerase kappa as an extender in translesion synthesis.Proc Natl Acad Sci U S A. 2002 Dec 10;99(25):16000-5. doi: 10.1073/pnas.252524999. Epub 2002 Nov 20. Proc Natl Acad Sci U S A. 2002. PMID: 12444249 Free PMC article.
-
Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions.Nature. 2000 Aug 31;406(6799):1015-9. doi: 10.1038/35023030. Nature. 2000. PMID: 10984059
-
DNA polymerase ζ in DNA replication and repair.Nucleic Acids Res. 2019 Sep 19;47(16):8348-8361. doi: 10.1093/nar/gkz705. Nucleic Acids Res. 2019. PMID: 31410467 Free PMC article. Review.
-
Mutagenesis in eukaryotes dependent on DNA polymerase zeta and Rev1p.Philos Trans R Soc Lond B Biol Sci. 2001 Jan 29;356(1405):41-6. doi: 10.1098/rstb.2000.0001. Philos Trans R Soc Lond B Biol Sci. 2001. PMID: 11205328 Free PMC article. Review.
Cited by
-
Proficient Replication of the Yeast Genome by a Viral DNA Polymerase.J Biol Chem. 2016 May 27;291(22):11698-705. doi: 10.1074/jbc.M116.728741. Epub 2016 Apr 12. J Biol Chem. 2016. PMID: 27072134 Free PMC article.
-
CDC7/DBF4 functions in the translesion synthesis branch of the RAD6 epistasis group in Saccharomyces cerevisiae.Genetics. 2004 Aug;167(4):1597-610. doi: 10.1534/genetics.103.021675. Genetics. 2004. PMID: 15342501 Free PMC article.
-
Rev1 overexpression accelerates N-methyl-N-nitrosourea (MNU)-induced thymic lymphoma by increasing mutagenesis.Cancer Sci. 2024 Jun;115(6):1808-1819. doi: 10.1111/cas.16159. Epub 2024 Apr 4. Cancer Sci. 2024. PMID: 38572512 Free PMC article.
-
Genetic instability in budding and fission yeast-sources and mechanisms.FEMS Microbiol Rev. 2015 Nov;39(6):917-67. doi: 10.1093/femsre/fuv028. Epub 2015 Jun 24. FEMS Microbiol Rev. 2015. PMID: 26109598 Free PMC article. Review.
-
Translesion Synthesis of 2'-Deoxyguanosine Lesions by Eukaryotic DNA Polymerases.Chem Res Toxicol. 2017 Jan 17;30(1):61-72. doi: 10.1021/acs.chemrestox.6b00285. Epub 2016 Nov 1. Chem Res Toxicol. 2017. PMID: 27760288 Free PMC article.
References
-
- Creighton, S., L. B. Bloom, and M. F. Goodman. 1995. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 262:232-256. - PubMed
-
- Goodman, M. F., S. Creighton, L. B. Bloom, and J. Petruska. 1993. Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol. 28:83-126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
