Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae
- PMID: 12556499
- PMCID: PMC141147
- DOI: 10.1128/MCB.23.4.1403-1417.2003
Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae
Abstract
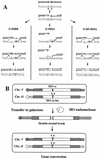
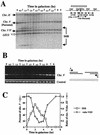
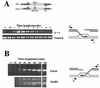
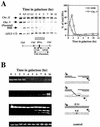
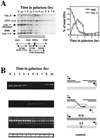
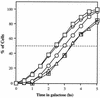
Recombination plays a central role in the repair of broken chromosomes in all eukaryotes. We carried out a systematic study of mitotic recombination. Using several assays, we established the chronological sequence of events necessary to repair a single double-strand break. Once a chromosome is broken, yeast cells become immediately committed to recombinational repair. Recombination is completed within an hour and exhibits two kinetic gaps. By using this kinetic framework we also characterized the role played by several proteins in the recombinational process. In the absence of Rad52, the broken chromosome ends, both 5' and 3', are rapidly degraded. This is not due to the inability to recombine, since the 3' single-stranded DNA ends are stable in a strain lacking donor sequences. Rad57 is required for two consecutive strand exchange reactions. Surprisingly, we found that the Srs2 helicase also plays an early positive role in the recombination process.
Figures




Similar articles
-
The role of DNA repair genes in recombination between repeated sequences in yeast.Genetics. 1995 Aug;140(4):1199-211. doi: 10.1093/genetics/140.4.1199. Genetics. 1995. PMID: 7498763 Free PMC article.
-
Genetic evidence for different RAD52-dependent intrachromosomal recombination pathways in Saccharomyces cerevisiae.Curr Genet. 1995 Mar;27(4):298-305. doi: 10.1007/BF00352096. Curr Genet. 1995. PMID: 7614550
-
Suppression of the double-strand-break-repair defect of the Saccharomyces cerevisiae rad57 mutant.Genetics. 2009 Apr;181(4):1195-206. doi: 10.1534/genetics.109.100842. Epub 2009 Feb 2. Genetics. 2009. PMID: 19189942 Free PMC article.
-
Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae.Microbiol Mol Biol Rev. 1999 Jun;63(2):349-404. doi: 10.1128/MMBR.63.2.349-404.1999. Microbiol Mol Biol Rev. 1999. PMID: 10357855 Free PMC article. Review.
-
Recombination proteins in yeast.Annu Rev Genet. 2004;38:233-71. doi: 10.1146/annurev.genet.38.072902.091500. Annu Rev Genet. 2004. PMID: 15568977 Review.
Cited by
-
A single Ho-induced double-strand break at the MAT locus is lethal in Candida glabrata.PLoS Genet. 2020 Oct 15;16(10):e1008627. doi: 10.1371/journal.pgen.1008627. eCollection 2020 Oct. PLoS Genet. 2020. PMID: 33057400 Free PMC article.
-
Extensive DNA end processing by exo1 and sgs1 inhibits break-induced replication.PLoS Genet. 2010 Jul 8;6(7):e1001007. doi: 10.1371/journal.pgen.1001007. PLoS Genet. 2010. PMID: 20628570 Free PMC article.
-
In vivo tracking of functionally tagged Rad51 unveils a robust strategy of homology search.Nat Struct Mol Biol. 2023 Oct;30(10):1582-1591. doi: 10.1038/s41594-023-01065-w. Epub 2023 Aug 21. Nat Struct Mol Biol. 2023. PMID: 37605042
-
Rad51 protein controls Rad52-mediated DNA annealing.J Biol Chem. 2008 May 23;283(21):14883-92. doi: 10.1074/jbc.M801097200. Epub 2008 Mar 12. J Biol Chem. 2008. PMID: 18337252 Free PMC article.
-
Mph1 and Mus81-Mms4 prevent aberrant processing of mitotic recombination intermediates.Mol Cell. 2013 Oct 10;52(1):63-74. doi: 10.1016/j.molcel.2013.09.007. Mol Cell. 2013. PMID: 24119400 Free PMC article.
References
-
- Allers, T., and M. Lichten. 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106:47-57. - PubMed
-
- Allers, T., and M. Lichten. 2001. Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol. Cell 8:225-231. - PubMed
-
- Fishman-Lobell, J., and J. E. Haber. 1992. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258:480-484. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
