Wobble modification differences and subcellular localization of tRNAs in Leishmania tarentolae: implication for tRNA sorting mechanism
- PMID: 12554666
- PMCID: PMC140750
- DOI: 10.1093/emboj/cdg066
Wobble modification differences and subcellular localization of tRNAs in Leishmania tarentolae: implication for tRNA sorting mechanism
Abstract
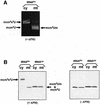
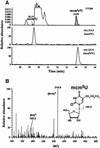
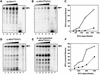
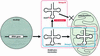
In Leishmania tarentolae, all mitochondrial tRNAs are encoded in the nuclear genome and imported from the cytosol. It is known that tRNA(Glu)(UUC) and tRNA(Gln)(UUG) are localized in both cytosol and mitochondria. We investigated structural differences between affinity-isolated cytosolic (cy) and mitochondrial (mt) tRNAs for glutamate and glutamine by mass spectrometry. A unique modification difference in both tRNAs was identified at the anticodon wobble position: cy tRNAs have 5-methoxycarbonylmethyl-2- thiouridine (mcm(5)s(2)U), whereas mt tRNAs have 5- methoxycarbonylmethyl-2'-O-methyluridine (mcm(5)Um). In addition, a trace portion (4%) of cy tRNAs was found to have 5-methoxycarbonylmethyluridine (mcm(5)U) at its wobble position, which could represent a common modification intermediate for both modified uridines in cy and mt tRNAs. We also isolated a trace amount of mitochondria-specific tRNA(Lys)(UUU) from the cytosol and found mcm(5)U at its wobble position, while its mitochondrial counterpart has mcm(5)Um. Mt tRNA(Lys) and in vitro transcribed tRNA(Glu) were imported much more efficiently into isolated mitochondria than the native cy tRNA(Glu) in an in vitro importation experiment, indicating that cytosol-specific 2-thiolation could play an inhibitory role in tRNA import into mitochondria.
Figures



Similar articles
-
Unusual usage of wobble modifications in mitochondrial tRNAs of the nematode Ascaris suum.FEBS Lett. 2005 May 23;579(13):2767-72. doi: 10.1016/j.febslet.2005.04.009. Epub 2005 Apr 20. FEBS Lett. 2005. PMID: 15907479
-
End processing precedes mitochondrial importation and editing of tRNAs in Leishmania tarentolae.J Biol Chem. 2000 Dec 1;275(48):37907-14. doi: 10.1074/jbc.M007838200. J Biol Chem. 2000. PMID: 10993905
-
A nuclear tRNA gene cluster in the protozoan Leishmania tarentolae and differential distribution of nuclear-encoded tRNAs between the cytosol and mitochondria.Mol Biochem Parasitol. 1994 May;65(1):23-37. doi: 10.1016/0166-6851(94)90112-0. Mol Biochem Parasitol. 1994. PMID: 7935626
-
Leishmania mitochondrial tRNA importers.Int J Biochem Cell Biol. 2008;40(12):2681-5. doi: 10.1016/j.biocel.2007.10.025. Epub 2007 Nov 20. Int J Biochem Cell Biol. 2008. PMID: 18061510 Review.
-
Sulfur Modifications of the Wobble U34 in tRNAs and their Intracellular Localization in Eukaryotic Cells.Biomolecules. 2017 Feb 18;7(1):17. doi: 10.3390/biom7010017. Biomolecules. 2017. PMID: 28218716 Free PMC article. Review.
Cited by
-
The T. brucei TRM5 methyltransferase plays an essential role in mitochondrial protein synthesis and function.RNA. 2013 May;19(5):649-58. doi: 10.1261/rna.036665.112. Epub 2013 Mar 21. RNA. 2013. PMID: 23520175 Free PMC article.
-
Thiolated tRNAs of Trypanosoma brucei are imported into mitochondria and dethiolated after import.J Biol Chem. 2009 Dec 25;284(52):36491-36499. doi: 10.1074/jbc.M109.064527. Epub 2009 Oct 29. J Biol Chem. 2009. PMID: 19875444 Free PMC article.
-
Biogenesis and functions of aminocarboxypropyluridine in tRNA.Nat Commun. 2019 Dec 5;10(1):5542. doi: 10.1038/s41467-019-13525-3. Nat Commun. 2019. PMID: 31804502 Free PMC article.
-
Methanothermobacter thermautotrophicus tRNA Gln confines the amidotransferase GatCAB to asparaginyl-tRNA Asn formation.J Mol Biol. 2008 Mar 28;377(3):845-53. doi: 10.1016/j.jmb.2008.01.064. Epub 2008 Jan 31. J Mol Biol. 2008. PMID: 18291416 Free PMC article.
-
tRNAs and proteins use the same import channel for translocation across the mitochondrial outer membrane of trypanosomes.Proc Natl Acad Sci U S A. 2017 Sep 12;114(37):E7679-E7687. doi: 10.1073/pnas.1711430114. Epub 2017 Aug 28. Proc Natl Acad Sci U S A. 2017. PMID: 28847952 Free PMC article.
References
-
- Adhya S., Ghosh,T., Das,A., Bera,S.K. and Mahapatra,S. (1997) Role of an RNA-binding protein in import of tRNA into Leishmania mitochondria. J. Biol. Chem., 272, 21396–21402. - PubMed
-
- Baczynskyj L., Biemann,K. and Hall,R.H. (1968) Sulfur-containing nucleoside from yeast transfer ribonucleic acid: 2-thio-5(or 6)-uridine acetic acid methyl ester. Science, 159, 1481–1483. - PubMed
-
- Braly P., Simpson,L. and Kretzer,F. (1974) Isolation of kinetoplast–mitochondrial complexes from Leishmania tarentolae. J. Protozool., 21, 782–790. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

