Heterochromatic sequences in a Drosophila whole-genome shotgun assembly
- PMID: 12537574
- PMCID: PMC151187
- DOI: 10.1186/gb-2002-3-12-research0085
Heterochromatic sequences in a Drosophila whole-genome shotgun assembly
Abstract
Background: Most eukaryotic genomes include a substantial repeat-rich fraction termed heterochromatin, which is concentrated in centric and telomeric regions. The repetitive nature of heterochromatic sequence makes it difficult to assemble and analyze. To better understand the heterochromatic component of the Drosophila melanogaster genome, we characterized and annotated portions of a whole-genome shotgun sequence assembly.
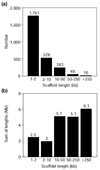
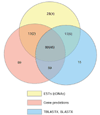
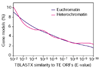
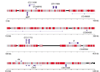
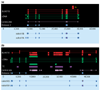
Results: WGS3, an improved whole-genome shotgun assembly, includes 20.7 Mb of draft-quality sequence not represented in the Release 3 sequence spanning the euchromatin. We annotated this sequence using the methods employed in the re-annotation of the Release 3 euchromatic sequence. This analysis predicted 297 protein-coding genes and six non-protein-coding genes, including known heterochromatic genes, and regions of similarity to known transposable elements. Bacterial artificial chromosome (BAC)-based fluorescence in situ hybridization analysis was used to correlate the genomic sequence with the cytogenetic map in order to refine the genomic definition of the centric heterochromatin; on the basis of our cytological definition, the annotated Release 3 euchromatic sequence extends into the centric heterochromatin on each chromosome arm.
Conclusions: Whole-genome shotgun assembly produced a reliable draft-quality sequence of a significant part of the Drosophila heterochromatin. Annotation of this sequence defined the intron-exon structures of 30 known protein-coding genes and 267 protein-coding gene models. The cytogenetic mapping suggests that an additional 150 predicted genes are located in heterochromatin at the base of the Release 3 euchromatic sequence. Our analysis suggests strategies for improving the sequence and annotation of the heterochromatic portions of the Drosophila and other complex genomes.
Figures


Similar articles
-
Sequence finishing and mapping of Drosophila melanogaster heterochromatin.Science. 2007 Jun 15;316(5831):1625-8. doi: 10.1126/science.1139816. Science. 2007. PMID: 17569867 Free PMC article.
-
The Release 6 reference sequence of the Drosophila melanogaster genome.Genome Res. 2015 Mar;25(3):445-58. doi: 10.1101/gr.185579.114. Epub 2015 Jan 14. Genome Res. 2015. PMID: 25589440 Free PMC article.
-
Y chromosome and other heterochromatic sequences of the Drosophila melanogaster genome: how far can we go?Genetica. 2003 Mar;117(2-3):227-37. doi: 10.1023/a:1022900313650. Genetica. 2003. PMID: 12723702
-
Euchromatic and heterochromatic domains at Drosophila telomeres.Biochem Cell Biol. 2005 Aug;83(4):477-85. doi: 10.1139/o05-053. Biochem Cell Biol. 2005. PMID: 16094451 Review.
-
A New Portrait of Constitutive Heterochromatin: Lessons from Drosophila melanogaster.Trends Genet. 2019 Sep;35(9):615-631. doi: 10.1016/j.tig.2019.06.002. Epub 2019 Jul 15. Trends Genet. 2019. PMID: 31320181 Review.
Cited by
-
α-Synuclein enhances histone H3 lysine-9 dimethylation and H3K9me2-dependent transcriptional responses.Sci Rep. 2016 Nov 3;6:36328. doi: 10.1038/srep36328. Sci Rep. 2016. PMID: 27808254 Free PMC article.
-
Genetic and bioinformatic analysis of 41C and the 2R heterochromatin of Drosophila melanogaster: a window on the heterochromatin-euchromatin junction.Genetics. 2004 Feb;166(2):807-22. doi: 10.1534/genetics.166.2.807. Genetics. 2004. PMID: 15020470 Free PMC article.
-
Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome.Genome Biol. 2006;7(11):R112. doi: 10.1186/gb-2006-7-11-r112. Genome Biol. 2006. PMID: 17134480 Free PMC article.
-
ReRep: computational detection of repetitive sequences in genome survey sequences (GSS).BMC Bioinformatics. 2008 Sep 9;9:366. doi: 10.1186/1471-2105-9-366. BMC Bioinformatics. 2008. PMID: 18782453 Free PMC article.
-
Satellite DNA-containing gigantic introns in a unique gene expression program during Drosophila spermatogenesis.PLoS Genet. 2019 May 9;15(5):e1008028. doi: 10.1371/journal.pgen.1008028. eCollection 2019 May. PLoS Genet. 2019. PMID: 31071079 Free PMC article.
References
-
- Heitz E. Das Heterochromatin der Moose. I Jahrb Wiss Botanik. 1928;69:762–818.
-
- John B. The biology of heterochromatin. In: Verma RS, editor. In Heterochromatin: Molecular and Structural Aspects. Cambridge: Cambridge University Press; 1988. pp. 1–147.
-
- Elgin SC, Workman JL. Chromosome and expression mechanisms: a year dominated by histone modifications, transitory and remembered. Curr Opin Genet Dev. 2002;12:127–129. - PubMed
-
- Weiler KS, Wakimoto BT. Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995;29:577–605. - PubMed
-
- Gatti M, Pimpinelli S. Functional elements in Drosophila melanogaster heterochromatin. Annu Rev Genet. 1992;26:239–275. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

