Replication of Carnation Italian ringspot virus defective interfering RNA in Saccharomyces cerevisiae
- PMID: 12525646
- PMCID: PMC140986
- DOI: 10.1128/jvi.77.3.2116-2123.2003
Replication of Carnation Italian ringspot virus defective interfering RNA in Saccharomyces cerevisiae
Abstract
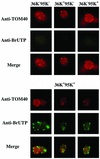
Two plasmids from which the sequences coding for the 36- and 95-kDa proteins of Carnation Italian ringspot virus (CIRV) could be transcribed in vivo in the yeast Saccharomyces cerevisiae under the control of the ADH1 promoter and terminator were constructed. The two proteins, which constitute the viral replicase, were correctly translated and integrated into membranes of the yeast cells. An additional plasmid was introduced in yeasts expressing the CIRV replicase, from which a defective interfering (DI) RNA (DI-7 RNA) could be transcribed under the control of the GAL1 promoter and terminated by the Tobacco ringspot virus satellite ribozyme, which cleaved 19 nucleotides downstream of the 3' end of DI RNA. The DI-7 RNA transcripts were amplified by the viral replicase as demonstrated by the restoration of the authentic 3' end, the requirement of a specific cis-acting signal at this terminus, the preferential accumulation of molecules with the authentic 5' terminus (AGAAA), the synthesis of head-to-tail dimers, the presence of negative strands, and the incorporation of 5-bromo-UTP. Additionally, transformation with a dimeric construct of DI-7 RNA led to the synthesis of monomers, mimicking the activity of the viral replicase in plant cells.
Figures




Similar articles
-
The p36 and p95 replicase proteins of Carnation Italian ringspot virus cooperate in stabilizing defective interfering RNA.J Gen Virol. 2004 Aug;85(Pt 8):2429-2433. doi: 10.1099/vir.0.80063-0. J Gen Virol. 2004. PMID: 15269385
-
Cytological analysis of Saccharomyces cerevisiae cells supporting cymbidium ringspot virus defective interfering RNA replication.J Gen Virol. 2006 Mar;87(Pt 3):705-714. doi: 10.1099/vir.0.81325-0. J Gen Virol. 2006. PMID: 16476994
-
Expression of tombusvirus open reading frames 1 and 2 is sufficient for the replication of defective interfering, but not satellite, RNA.J Gen Virol. 2004 Oct;85(Pt 10):3115-3122. doi: 10.1099/vir.0.80296-0. J Gen Virol. 2004. PMID: 15448375
-
Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication.Virology. 2006 Jan 5;344(1):211-20. doi: 10.1016/j.virol.2005.09.017. Virology. 2006. PMID: 16364751 Review.
-
Origin and replication of defective interfering particles.Curr Top Microbiol Immunol. 1981;93:151-207. doi: 10.1007/978-3-642-68123-3_7. Curr Top Microbiol Immunol. 1981. PMID: 7026180 Review. No abstract available.
Cited by
-
Impediometric Electrochemical Sensor Based on The Inspiration of Carnation Italian Ringspot Virus Structure to Detect an Attommolar of miR.Sci Rep. 2020 Jun 15;10(1):9645. doi: 10.1038/s41598-020-66393-z. Sci Rep. 2020. PMID: 32541792 Free PMC article.
-
Expression of the Cymbidium ringspot virus 33-kilodalton protein in Saccharomyces cerevisiae and molecular dissection of the peroxisomal targeting signal.J Virol. 2004 May;78(9):4744-52. doi: 10.1128/jvi.78.9.4744-4752.2004. J Virol. 2004. PMID: 15078956 Free PMC article.
-
Susceptibility Genes to Plant Viruses.Viruses. 2018 Sep 10;10(9):484. doi: 10.3390/v10090484. Viruses. 2018. PMID: 30201857 Free PMC article. Review.
-
Yeast and the AIDS virus: the odd couple.J Biomed Biotechnol. 2012;2012:549020. doi: 10.1155/2012/549020. Epub 2012 Jun 17. J Biomed Biotechnol. 2012. PMID: 22778552 Free PMC article. Review.
-
Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication.J Virol. 2006 Mar;80(5):2162-9. doi: 10.1128/JVI.80.5.2162-2169.2006. J Virol. 2006. PMID: 16474124 Free PMC article.
References
-
- Ammerer, G. 1983. Expression of genes in yeast using ADC1 promoter. Methods Enzymol. 101:192-201. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
-
- Baker, K. P., A. Schaniel, D. Vestweber, and G. Schatz. 1990. A yeast mitochondrial outer membrane protein essential for protein import and cell viability. Nature 348:605-609. - PubMed
-
- Been, M. D. 1994. cis- and trans-acting ribozymes from a human pathogen, hepatitis delta virus. Trends Biochem. Sci. 19:251-256. - PubMed
-
- Berben, G., J. Dumont, V. Gilliquet, P. A. Bolle, and F. Hilger. 1991. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast 7:475-477. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

