Human immunodeficiency virus type 1 nucleocapsid zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA
- PMID: 12502862
- PMCID: PMC140799
- DOI: 10.1128/jvi.77.2.1469-1480.2003
Human immunodeficiency virus type 1 nucleocapsid zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA
Abstract
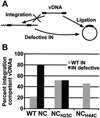
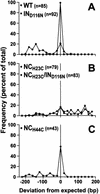
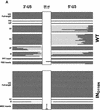
Human immunodeficiency virus type 1 (HIV-1) containing mutations in the nucleocapsid (NC) Zn(2+) finger domains have greatly reduced infectivity, even though genome packaging is largely unaffected in certain cases. To examine replication defects, viral DNA (vDNA) was isolated from cells infected with viruses containing His-to-Cys changes in their Zn(2+) fingers (NC(H23C) and NC(H44C)), an integrase mutant (IN(D116N)), a double mutant (NC(H23C)/IN(D116N)), or wild-type HIV-1. In vitro assays have established potential roles for NC in reverse transcription and integration. In vivo results for these processes were obtained by quantitative PCR, cloning of PCR products, and comparison of the quantity and composition of vDNA generated at discrete points during reverse transcription. Quantitative analysis of the reverse transcription intermediates for these species strongly suggests decreased stability of the DNA produced. Both Zn(2+) finger mutants appear to be defective in DNA synthesis, with the minus- and plus-strand transfer processes being affected while interior portions of the vDNA remain more intact. Sequences obtained from PCR amplification and cloning of 2-LTR circle junction fragments revealed that the NC mutants had a phenotype similar to the IN mutant; removal of the terminal CA dinucleotides necessary for integration of the vDNA is disabled by the NC mutations. Thus, the loss of infectivity in these NC mutants in vivo appears to result from defective reverse transcription and integration processes stemming from decreased protection of the full-length vDNA. Finally, these results indicate that the chaperone activity of NC extends from the management of viral RNA through to the full-length vDNA.
Figures



Similar articles
-
Human immunodeficiency virus type 1 nucleocapsid zinc-finger mutations cause defects in reverse transcription and integration.Virology. 2006 Sep 15;353(1):41-51. doi: 10.1016/j.virol.2006.05.014. Epub 2006 Jun 19. Virology. 2006. PMID: 16784767
-
Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences.Virology. 1999 Mar 30;256(1):92-104. doi: 10.1006/viro.1999.9629. Virology. 1999. PMID: 10087230
-
Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer.J Virol. 2000 Oct;74(19):8980-8. doi: 10.1128/jvi.74.19.8980-8988.2000. J Virol. 2000. PMID: 10982342 Free PMC article.
-
Nucleocapsid protein function in early infection processes.Virus Res. 2008 Jun;134(1-2):39-63. doi: 10.1016/j.virusres.2007.12.006. Epub 2008 Feb 14. Virus Res. 2008. PMID: 18279991 Free PMC article. Review.
-
Site-specific integration of retroviral DNA in human cells using fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc-finger protein E2C.Methods. 2009 Apr;47(4):269-76. doi: 10.1016/j.ymeth.2009.01.001. Epub 2009 Jan 30. Methods. 2009. PMID: 19186211 Free PMC article. Review.
Cited by
-
TNPO3 protects HIV-1 replication from CPSF6-mediated capsid stabilization in the host cell cytoplasm.Retrovirology. 2013 Feb 15;10:20. doi: 10.1186/1742-4690-10-20. Retrovirology. 2013. PMID: 23414560 Free PMC article.
-
Single-molecule spectroscopic study of dynamic nanoscale DNA bending behavior of HIV-1 nucleocapsid protein.J Phys Chem B. 2013 Apr 25;117(16):4183-96. doi: 10.1021/jp3018259. Epub 2012 May 16. J Phys Chem B. 2013. PMID: 22591315 Free PMC article.
-
Proteomic analysis of early HIV-1 nucleoprotein complexes.J Proteome Res. 2013 Feb 1;12(2):559-72. doi: 10.1021/pr300869h. Epub 2013 Jan 16. J Proteome Res. 2013. PMID: 23282062 Free PMC article.
-
Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides.Nucleic Acids Res. 2006 Jan 24;34(2):472-84. doi: 10.1093/nar/gkj442. Print 2006. Nucleic Acids Res. 2006. PMID: 16434700 Free PMC article.
-
Heterologous human immunodeficiency virus type 1 lentiviral vectors packaging a simian immunodeficiency virus-derived genome display a specific postentry transduction defect in dendritic cells.J Virol. 2003 Sep;77(17):9295-304. doi: 10.1128/jvi.77.17.9295-9304.2003. J Virol. 2003. PMID: 12915545 Free PMC article.
References
-
- Berg, J. M. 1986. Potential metal-binding domains in nucleic acid binding proteins. Science 232:485-487. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

