Biochemical and genetic studies of the VPg uridylylation reaction catalyzed by the RNA polymerase of poliovirus
- PMID: 12502805
- PMCID: PMC140777
- DOI: 10.1128/jvi.77.2.891-904.2003
Biochemical and genetic studies of the VPg uridylylation reaction catalyzed by the RNA polymerase of poliovirus
Abstract
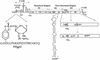
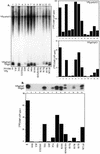
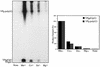
The first step in poliovirus (PV) RNA synthesis is the covalent linkage of UMP to the terminal protein VPg. This reaction can be studied in vitro with two different assays. The simpler assay is based on a poly(A) template and requires synthetic VPg, purified RNA polymerase 3D(pol), UTP, and a divalent cation. The other assay uses specific viral sequences [cre(2C)] as a template for VPg uridylylation and requires the addition of proteinase 3CD(pro). Using one or both of these assays, we analyzed the VPg specificities and metal requirements of the uridylylation reactions. We determined the effects of single and double amino acid substitutions in VPg on the abilities of the peptides to serve as substrates for 3D(pol). Mutations in VPg, which interfered with uridylylation in vitro, were found to abolish viral growth. A chimeric PV containing the VPg of human rhinovirus 14 (HRV14) was viable, but substitutions of HRV2 and HRV89 VPgs for PV VPg were lethal. Of the three rhinoviral VPgs tested, only the HRV14 peptide was found to function as a substrate for PV1(M) 3D(pol) in vitro. We also examined the metal specificity of the VPg uridylylation reaction on a poly(A) template. Our results show a strong preference of the RNA polymerase for Mn(2+) as a cofactor compared to Mg(2+) or other divalent cations.
Figures





Similar articles
-
Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg.J Virol. 2000 Nov;74(22):10359-70. doi: 10.1128/jvi.74.22.10359-10370.2000. J Virol. 2000. PMID: 11044080 Free PMC article.
-
Studies on the attenuation phenotype of polio vaccines: poliovirus RNA polymerase derived from Sabin type 1 sequence is temperature sensitive in the uridylylation of VPg.Virology. 2000 Jun 20;272(1):72-84. doi: 10.1006/viro.2000.0354. Virology. 2000. PMID: 10873750
-
Viability of poliovirus/rhinovirus VPg chimeric viruses and identification of an amino acid residue in the VPg gene critical for viral RNA replication.J Virol. 2003 Jul;77(13):7434-43. doi: 10.1128/jvi.77.13.7434-7443.2003. J Virol. 2003. PMID: 12805442 Free PMC article.
-
Uridylylation of the genome-linked protein of poliovirus in vitro is dependent upon an endogenous RNA template.Virus Res. 1987 Sep;8(3):193-204. doi: 10.1016/0168-1702(87)90015-3. Virus Res. 1987. PMID: 2825442 Review.
-
Initiation of protein-primed picornavirus RNA synthesis.Virus Res. 2015 Aug 3;206:12-26. doi: 10.1016/j.virusres.2014.12.028. Epub 2015 Jan 12. Virus Res. 2015. PMID: 25592245 Free PMC article. Review.
Cited by
-
Viral polymerases.Adv Exp Med Biol. 2012;726:267-304. doi: 10.1007/978-1-4614-0980-9_12. Adv Exp Med Biol. 2012. PMID: 22297518 Free PMC article. Review.
-
Crystal structure of enterovirus 71 RNA-dependent RNA polymerase complexed with its protein primer VPg: implication for a trans mechanism of VPg uridylylation.J Virol. 2013 May;87(10):5755-68. doi: 10.1128/JVI.02733-12. Epub 2013 Mar 13. J Virol. 2013. PMID: 23487447 Free PMC article.
-
NMR solution structure of poliovirus uridylyated peptide linked to the genome (VPgpU).Peptides. 2010 Aug;31(8):1441-8. doi: 10.1016/j.peptides.2010.04.021. Epub 2010 May 2. Peptides. 2010. PMID: 20441784 Free PMC article.
-
Mutational Disruption of cis-Acting Replication Element 2C in Coxsackievirus B3 Leads to 5'-Terminal Genomic Deletions.J Virol. 2015 Dec;89(23):11761-72. doi: 10.1128/JVI.01308-15. Epub 2015 Sep 9. J Virol. 2015. PMID: 26355088 Free PMC article.
-
Factors required for the Uridylylation of the foot-and-mouth disease virus 3B1, 3B2, and 3B3 peptides by the RNA-dependent RNA polymerase (3Dpol) in vitro.J Virol. 2005 Jun;79(12):7698-706. doi: 10.1128/JVI.79.12.7698-7706.2005. J Virol. 2005. PMID: 15919922 Free PMC article.
References
-
- Agol, V. I., A. V. Paul, and E. Wimmer. 1999. Paradoxes of the replication of picornaviral genomes. Virus Res. 62:129-147. - PubMed
-
- Ambros, V., and D. Baltimore. 1978. Protein is linked to the 5′ end of poliovirus RNA by a phosphodiester linkage to tyrosine. J. Biol. Chem. 253:5263-5266. - PubMed
-
- Arnold, J. J., S. K. B. Ghosh, and C. E. Cameron. 1999. Poliovirus RNA-dependent RNA polymerase (3Dpol). Divalent cation modulation of primer, template, and nucleotide selection. J. Biol. Chem. 274:37060-37069. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

