Structural basis and prediction of substrate specificity in protein serine/threonine kinases
- PMID: 12502784
- PMCID: PMC140887
- DOI: 10.1073/pnas.0134224100
Structural basis and prediction of substrate specificity in protein serine/threonine kinases
Abstract
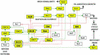
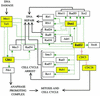
The large number of protein kinases makes it impractical to determine their specificities and substrates experimentally. Using the available crystal structures, molecular modeling, and sequence analyses of kinases and substrates, we developed a set of rules governing the binding of a heptapeptide substrate motif (surrounding the phosphorylation site) to the kinase and implemented these rules in a web-interfaced program for automated prediction of optimal substrate peptides, taking only the amino acid sequence of a protein kinase as input. We show the utility of the method by analyzing yeast cell cycle control and DNA damage checkpoint pathways. Our method is the only available predictive method generally applicable for identifying possible substrate proteins for protein serinethreonine kinases and helps in silico construction of signaling pathways. The accuracy of prediction is comparable to the accuracy of data from systematic large-scale experimental approaches.
Figures
Similar articles
-
Identification of substrates for Ser/Thr kinases using residue-based statistical pair potentials.Bioinformatics. 2010 Jan 15;26(2):189-97. doi: 10.1093/bioinformatics/btp633. Epub 2009 Nov 12. Bioinformatics. 2010. PMID: 19910306
-
Phosphorylation target site specificity for AGC kinases DMPK E and Lats2.J Cell Biochem. 2012 Jun;113(6):2126-35. doi: 10.1002/jcb.24086. J Cell Biochem. 2012. PMID: 22492269
-
A computational analysis of substrate binding strength by phosphorylase kinase and protein kinase A.J Mol Recognit. 2002 Mar-Apr;15(2):104-11. doi: 10.1002/jmr.563. J Mol Recognit. 2002. PMID: 11954055
-
Molecular mechanisms controlling the localisation of protein kinase A.Curr Genet. 2002 Jul;41(4):199-207. doi: 10.1007/s00294-002-0308-9. Epub 2002 Jun 27. Curr Genet. 2002. PMID: 12172960 Review.
-
Signalling specificity of Ser/Thr protein kinases through docking-site-mediated interactions.Biochem J. 2003 May 15;372(Pt 1):1-13. doi: 10.1042/BJ20021641. Biochem J. 2003. PMID: 12600273 Free PMC article. Review.
Cited by
-
Prediction of human disease-associated phosphorylation sites with combined feature selection approach and support vector machine.IET Syst Biol. 2015 Aug;9(4):155-63. doi: 10.1049/iet-syb.2014.0051. IET Syst Biol. 2015. PMID: 26243832 Free PMC article.
-
The kinase Grk2 regulates Nedd4/Nedd4-2-dependent control of epithelial Na+ channels.Proc Natl Acad Sci U S A. 2004 Aug 10;101(32):11886-90. doi: 10.1073/pnas.0402178101. Epub 2004 Jul 29. Proc Natl Acad Sci U S A. 2004. PMID: 15284439 Free PMC article.
-
Probabilistic approach to predicting substrate specificity of methyltransferases.PLoS Comput Biol. 2014 Mar 20;10(3):e1003514. doi: 10.1371/journal.pcbi.1003514. eCollection 2014 Mar. PLoS Comput Biol. 2014. PMID: 24651469 Free PMC article.
-
Discovery of phosphorylation motif mixtures in phosphoproteomics data.Bioinformatics. 2009 Jan 1;25(1):14-21. doi: 10.1093/bioinformatics/btn569. Epub 2008 Nov 7. Bioinformatics. 2009. PMID: 18996944 Free PMC article.
-
The Predikin webserver: improved prediction of protein kinase peptide specificity using structural information.Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W286-90. doi: 10.1093/nar/gkn279. Epub 2008 May 13. Nucleic Acids Res. 2008. PMID: 18477637 Free PMC article.
References
-
- Songyang Z, Blechner S, Hoagland N, Hoekstra M F, Piwnica-Worms H, Cantley L C. Curr Biol. 1994;4:973–982. - PubMed
-
- Hardie D G. Protein Phosphorylation. Oxford: Oxford Univ. Press; 1999.
-
- Bishop A C, Ubersax J A, Petsch D T, Matheos D P, Gray N S, Blethrow J, Shimizu E, Tsien J Z, Schultz P G, Rose M D, et al. Nature. 2000;407:395–401. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

