Monitoring mis-acylated tRNA suppression efficiency in mammalian cells via EGFP fluorescence recovery
- PMID: 12466560
- PMCID: PMC137983
- DOI: 10.1093/nar/gnf128
Monitoring mis-acylated tRNA suppression efficiency in mammalian cells via EGFP fluorescence recovery
Abstract
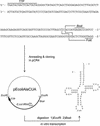
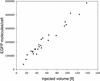
A reporter assay was developed to detect and quantify nonsense codon suppression by chemically aminoacylated tRNAs in mammalian cells. It is based on the cellular expression of the enhanced green fluorescent protein (EGFP) as a reporter for the site-specific amino acid incorporation in its sequence using an orthogonal suppressor tRNA derived from Escherichia coli. Suppression of an engineered amber codon at position 64 in the EGFP run-off transcript could be achieved by the incorporation of a leucine via an in vitro aminoacylated suppressor tRNA. Microinjection of defined amounts of mutagenized EGFP mRNA and suppressor tRNA into individual cells allowed us to accurately determine suppression efficiencies by measuring the EGFP fluorescence intensity in individual cells using laser-scanning confocal microscopy. Control experiments showed the absence of natural suppression or aminoacylation of the synthetic tRNA by endogenous aminoacyl-tRNA synthetases. This reporter assay opens the way for the optimization of essential experimental parameters for expanding the scope of the suppressor tRNA technology to different cell types.
Figures


Similar articles
-
Complete set of orthogonal 21st aminoacyl-tRNA synthetase-amber, ochre and opal suppressor tRNA pairs: concomitant suppression of three different termination codons in an mRNA in mammalian cells.Nucleic Acids Res. 2004 Dec 1;32(21):6200-11. doi: 10.1093/nar/gkh959. Print 2004. Nucleic Acids Res. 2004. PMID: 15576346 Free PMC article.
-
Downregulation of eRF1 by RNA interference increases mis-acylated tRNA suppression efficiency in human cells.Protein Eng Des Sel. 2004 Dec;17(12):821-7. doi: 10.1093/protein/gzh096. Epub 2005 Feb 16. Protein Eng Des Sel. 2004. PMID: 15716307
-
Engineering of an orthogonal aminoacyl-tRNA synthetase for efficient incorporation of the non-natural amino acid O-methyl-L-tyrosine using fluorescence-based bacterial cell sorting.J Mol Biol. 2010 Nov 19;404(1):70-87. doi: 10.1016/j.jmb.2010.09.001. Epub 2010 Sep 15. J Mol Biol. 2010. PMID: 20837025
-
Engineering the Genetic Code in Cells and Animals: Biological Considerations and Impacts.Acc Chem Res. 2017 Nov 21;50(11):2767-2775. doi: 10.1021/acs.accounts.7b00376. Epub 2017 Oct 6. Acc Chem Res. 2017. PMID: 28984438 Free PMC article. Review.
-
Plasticity and Constraints of tRNA Aminoacylation Define Directed Evolution of Aminoacyl-tRNA Synthetases.Int J Mol Sci. 2019 May 9;20(9):2294. doi: 10.3390/ijms20092294. Int J Mol Sci. 2019. PMID: 31075874 Free PMC article. Review.
Cited by
-
Incorporation of nonstandard amino acids into proteins: principles and applications.World J Microbiol Biotechnol. 2020 Apr 8;36(4):60. doi: 10.1007/s11274-020-02837-y. World J Microbiol Biotechnol. 2020. PMID: 32266578 Review.
-
Stress Response and Adaptation Mediated by Amino Acid Misincorporation during Protein Synthesis.Adv Nutr. 2016 Jul 15;7(4):773S-9S. doi: 10.3945/an.115.010991. Print 2016 Jul. Adv Nutr. 2016. PMID: 27422514 Free PMC article. Review.
-
Sugar nucleotide pools of Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major.Eukaryot Cell. 2007 Aug;6(8):1450-63. doi: 10.1128/EC.00175-07. Epub 2007 Jun 8. Eukaryot Cell. 2007. PMID: 17557881 Free PMC article.
-
Complete set of orthogonal 21st aminoacyl-tRNA synthetase-amber, ochre and opal suppressor tRNA pairs: concomitant suppression of three different termination codons in an mRNA in mammalian cells.Nucleic Acids Res. 2004 Dec 1;32(21):6200-11. doi: 10.1093/nar/gkh959. Print 2004. Nucleic Acids Res. 2004. PMID: 15576346 Free PMC article.
-
Mistranslation of the genetic code.FEBS Lett. 2014 Nov 28;588(23):4305-10. doi: 10.1016/j.febslet.2014.08.035. Epub 2014 Sep 16. FEBS Lett. 2014. PMID: 25220850 Free PMC article. Review.
References
-
- Gallivan J.P., Lester,H.A. and Dougherty,D.A. (1997) Site-specific incorporation of biotinylated amino acids to identify surface-exposed residues in integral membrane proteins. Chem. Biol., 4, 739–749. - PubMed
-
- Mamaev S.V., Laikhter,A.L., Arslan,T. and Hecht,S.M. (1996) Firefly luciferase: alteration of the color of emitted light resulting from substitutions at position 286. J. Am. Chem. Soc., 118, 7243–7244.
-
- Mendel D., Cornish,V.W. and Schultz,P.G. (1995) Site-directed mutagenesis with an expanded genetic code. Annu. Rev. Biophys. Biomol. Struct., 24, 435–462. - PubMed
-
- Turcatti G., Nemeth,K., Edgerton,M.D., Meseth,U., Talabot,F., Peitsch,M., Knowles,J., Vogel,H. and Chollet,A. (1996) Probing the structure and function of the tachykinin neurokinin-2 receptor through biosynthetic incorporation of fluorescent amino acids at specific sites. J. Biol. Chem., 271, 19991–19998. - PubMed
-
- Cohen B.E., McAnaney,T.B., Park,E.S., Jan,Y.N., Boxer,S.G. and Jan,L.Y. (2002) Probing protein electrostatics with a synthetic fluorescent amino acid. Science, 296, 1700–1703. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

