Domain regulation of imprinting cluster in Kip2/Lit1 subdomain on mouse chromosome 7F4/F5: large-scale DNA methylation analysis reveals that DMR-Lit1 is a putative imprinting control region
- PMID: 12466290
- PMCID: PMC187562
- DOI: 10.1101/gr.110702
Domain regulation of imprinting cluster in Kip2/Lit1 subdomain on mouse chromosome 7F4/F5: large-scale DNA methylation analysis reveals that DMR-Lit1 is a putative imprinting control region
Erratum in
-
Domain regulation of imprinting cluster in Kip2/Lit1 subdomain on mouse chromosome 7F4/F5: large-scale DNA methylation analysis reveals that DMR-Lit1 is a putative imprinting control region.Genome Res. 2004 Sep;14(9):1820. Genome Res. 2004. PMID: 15378833 Free PMC article. No abstract available.
Abstract
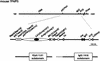
Mouse chromosome 7F4/F5, where the imprinting domain is located, is syntenic to human 11p15.5, the locus for Beckwith-Wiedemann syndrome. The domain is thought to consist of the two subdomains Kip2 (p57(kip2))/Lit1 and Igf2/H19. Because DNA methylation is believed to be a key factor in genomic imprinting, we performed large-scale DNA methylation analysis to identify the cis-element crucial for the regulation of the Kip2/Lit1 subdomain. Ten CpG islands (CGIs) were found, and these were located at the promoter sites, upstream of genes, and within intergenic regions. Bisulphite sequencing revealed that CGIs 4, 5, 8, and 10 were differentially methylated regions (DMRs). CGIs 4, 5, and 10 were methylated paternally in somatic tissues but not in germ cells. CGI8 was methylated in oocyte and maternally in somatic tissues during development. Parental-specific DNase I hypersensitive sites (HSSs) were found near CGI8. These data indicate that CGI8, called DMR-Lit1, is not only the region for gametic methylation but might also be the imprinting control region (ICR) of the subdomain.
Figures

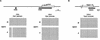
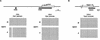




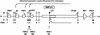
Similar articles
-
Silencing of imprinted CDKN1C gene expression is associated with loss of CpG and histone H3 lysine 9 methylation at DMR-LIT1 in esophageal cancer.Oncogene. 2004 May 27;23(25):4380-8. doi: 10.1038/sj.onc.1207576. Oncogene. 2004. PMID: 15007390
-
Loss of CpG methylation is strongly correlated with loss of histone H3 lysine 9 methylation at DMR-LIT1 in patients with Beckwith-Wiedemann syndrome.Am J Hum Genet. 2003 Oct;73(4):948-56. doi: 10.1086/378595. Epub 2003 Aug 29. Am J Hum Genet. 2003. PMID: 12949703 Free PMC article.
-
Targeted disruption of the human LIT1 locus defines a putative imprinting control element playing an essential role in Beckwith-Wiedemann syndrome.Hum Mol Genet. 2000 Sep 1;9(14):2075-83. doi: 10.1093/hmg/9.14.2075. Hum Mol Genet. 2000. PMID: 10958646
-
Genomic imprinting and chromatin insulation in Beckwith-Wiedemann syndrome.Mol Biotechnol. 1999 Apr;11(2):159-73. doi: 10.1007/BF02915809. Mol Biotechnol. 1999. PMID: 10464770 Review.
-
Methylated DNA sequences in genomic imprinting.Crit Rev Eukaryot Gene Expr. 2000;10(3-4):241-57. doi: 10.1615/critreveukargeneexpr.v10.i3-4.30. Crit Rev Eukaryot Gene Expr. 2000. PMID: 11272467 Review.
Cited by
-
MyoD regulates p57kip2 expression by interacting with a distant cis-element and modifying a higher order chromatin structure.Nucleic Acids Res. 2012 Sep 1;40(17):8266-75. doi: 10.1093/nar/gks619. Epub 2012 Jun 26. Nucleic Acids Res. 2012. PMID: 22740650 Free PMC article.
-
Methylation is maintained specifically at imprinting control regions but not other DMRs associated with imprinted genes in mice bearing a mutation in the Dnmt1 intrinsically disordered domain.Front Cell Dev Biol. 2023 Aug 4;11:1192789. doi: 10.3389/fcell.2023.1192789. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37601113 Free PMC article.
-
Asymmetric DNA methylation of CpG dyads is a feature of secondary DMRs associated with the Dlk1/Gtl2 imprinting cluster in mouse.Epigenetics Chromatin. 2017 Jun 21;10:31. doi: 10.1186/s13072-017-0138-0. eCollection 2017. Epigenetics Chromatin. 2017. PMID: 28649282 Free PMC article.
-
Drug-induced loss of imprinting revealed using bioluminescent reporters of Cdkn1c.Sci Rep. 2023 Apr 6;13(1):5626. doi: 10.1038/s41598-023-32747-6. Sci Rep. 2023. PMID: 37024615 Free PMC article.
-
MIRA-SNuPE, a quantitative, multiplex method for measuring allele-specific DNA methylation.Epigenetics. 2011 Feb;6(2):212-23. doi: 10.4161/epi.6.2.13699. Epub 2011 Feb 1. Epigenetics. 2011. PMID: 20948294 Free PMC article.
References
-
- Avner P, Heard E. X-chromosome inactivation: Counting, choice and initiation. Nat Rev Genet. 2001;2:59–67. - PubMed
-
- Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10:687–692. - PubMed
-
- Ben-Porath I, Cedar H. Imprinting: Focusing on the center. Curr Opin Genet Dev. 2000;10:550–554. - PubMed
-
- Bourchis D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
