Divergent subunit interactions among fungal mRNA 5'-capping machineries
- PMID: 12455993
- PMCID: PMC118010
- DOI: 10.1128/EC.1.3.448-457.2002
Divergent subunit interactions among fungal mRNA 5'-capping machineries
Abstract
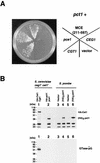
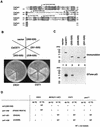
The Saccharomyces cerevisiae mRNA capping enzyme consists of two subunits: an RNA 5'-triphosphatase (RTPase) and GTP::mRNA guanylyltransferase (GTase). The GTase subunit (Ceg1) binds to the phosphorylated carboxyl-terminal domain of the largest subunit (CTD-P) of RNA polymerase II (pol II), coupling capping with transcription. Ceg1 bound to the CTD-P is inactive unless allosterically activated by interaction with the RTPase subunit (Cet1). For purposes of comparison, we characterize here the related GTases and RTPases from the yeasts Schizosaccharomyces pombe and Candida albicans. Surprisingly, the S. pombe capping enzyme subunits do not interact with each other. Both can independently interact with CTD-P of pol II, and the GTase is not repressed by CTD-P binding. The S. pombe RTPase gene (pct1+) is essential for viability. Pct1 can replace the S. cerevisiae RTPase when GTase activity is supplied by the S. pombe or mouse enzymes but not by the S. cerevisiae GTase. The C. albicans capping enzyme subunits do interact with each other. However, this interaction is not essential in vivo. Our results reveal an unexpected diversity among the fungal capping machineries.
Figures

Similar articles
-
An essential function of Saccharomyces cerevisiae RNA triphosphatase Cet1 is to stabilize RNA guanylyltransferase Ceg1 against thermal inactivation.J Biol Chem. 2001 Sep 28;276(39):36116-24. doi: 10.1074/jbc.M105856200. Epub 2001 Jul 19. J Biol Chem. 2001. PMID: 11463793
-
Isolation and characterization of the Candida albicans gene for mRNA 5'-triphosphatase: association of mRNA 5'-triphosphatase and mRNA 5'-guanylyltransferase activities is essential for the function of mRNA 5'-capping enzyme in vivo.FEBS Lett. 1998 Sep 11;435(1):49-54. doi: 10.1016/s0014-5793(98)01037-0. FEBS Lett. 1998. PMID: 9755857
-
Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5.J Biol Chem. 2002 May 31;277(22):19639-48. doi: 10.1074/jbc.M200015200. Epub 2002 Mar 13. J Biol Chem. 2002. PMID: 11893740
-
DNA polymerase II, the epsilon polymerase of Saccharomyces cerevisiae.Prog Nucleic Acid Res Mol Biol. 1993;46:93-120. doi: 10.1016/s0079-6603(08)61019-3. Prog Nucleic Acid Res Mol Biol. 1993. PMID: 8234788 Review. No abstract available.
-
Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human.Nucleic Acids Res. 2001 Jul 1;29(13):2675-90. doi: 10.1093/nar/29.13.2675. Nucleic Acids Res. 2001. PMID: 11433012 Free PMC article. Review.
Cited by
-
Deletion of individual mRNA capping genes is unexpectedly not lethal to Candida albicans and results in modified mRNA cap structures.Eukaryot Cell. 2002 Dec;1(6):1010-20. doi: 10.1128/EC.1.6.1010-1020.2002. Eukaryot Cell. 2002. PMID: 12477801 Free PMC article.
-
Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA capping apparatus.Structure. 2010 Feb 10;18(2):216-27. doi: 10.1016/j.str.2009.12.009. Structure. 2010. PMID: 20159466 Free PMC article.
-
Cyclin-dependent kinase 9 (Cdk9) of fission yeast is activated by the CDK-activating kinase Csk1, overlaps functionally with the TFIIH-associated kinase Mcs6, and associates with the mRNA cap methyltransferase Pcm1 in vivo.Mol Cell Biol. 2006 Feb;26(3):777-88. doi: 10.1128/MCB.26.3.777-788.2006. Mol Cell Biol. 2006. PMID: 16428435 Free PMC article.
-
Enzymology of RNA cap synthesis.Wiley Interdiscip Rev RNA. 2010 Jul-Aug;1(1):152-72. doi: 10.1002/wrna.19. Epub 2010 May 25. Wiley Interdiscip Rev RNA. 2010. PMID: 21956912 Free PMC article. Review.
-
Defenders of the Transcriptome: Guard Protein-Mediated mRNA Quality Control in Saccharomyces cerevisiae.Int J Mol Sci. 2024 Sep 24;25(19):10241. doi: 10.3390/ijms251910241. Int J Mol Sci. 2024. PMID: 39408571 Free PMC article. Review.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1991. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
-
- Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1988. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

