RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome
- PMID: 12438618
- PMCID: PMC136701
- DOI: 10.1128/jvi.76.24.12925-12933.2002
RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome
Abstract
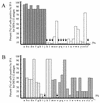
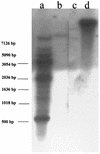
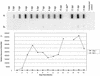
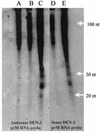
Double-stranded RNA (dsRNA) initiates cellular posttranscriptional responses that are collectively called RNA silencing in a number of different organisms, including plants, nematodes, and fruit flies. In plants, RNA silencing has been associated with protection from virus infection. In this study, we demonstrate that dsRNA-mediated interference also can act as a viral defense mechanism in mosquito cells. C6/36 (Aedes albopictus) cells were stably transformed with a plasmid designed to transcribe an inverted-repeat RNA (irRNA) derived from the genome of dengue virus type 2 (DEN-2) capable of forming dsRNA. Clonal cell lines were selected with an antibiotic resistance marker and challenged with DEN-2. The cell lines were classified as either susceptible or resistant to virus replication, based on the percentage of cells expressing DEN-2 envelope (E) antigen 7 days after challenge. Eight out of 18 (44%) cell lines designed to express irRNA were resistant to DEN-2 challenge, with more than 95% of the cells showing no DEN-2 antigen accumulation. One of the DEN-2-resistant cell lines, FB 9.1, was further characterized. DEN-2 genome RNA failed to accumulate in FB 9.1 cells after challenge. Northern blot hybridization detected transcripts containing transgene sequences of both sense and antisense polarity, suggesting that DEN-2-specific dsRNA was present in the cells. In addition, a class of small RNAs 21 to 25 nucleotides in length was detected that specifically hybridized to labeled sense or antisense DEN-2 RNA derived from the target region of the genome. These observations were consistent with RNA silencing as the mechanism of resistance to DEN-2 in transformed mosquito cells.
Figures




Similar articles
-
Pathogen-derived resistance to dengue type 2 virus in mosquito cells by expression of the premembrane coding region of the viral genome.J Virol. 1996 Apr;70(4):2132-7. doi: 10.1128/JVI.70.4.2132-2137.1996. J Virol. 1996. PMID: 8642634 Free PMC article.
-
Inhibition of viral gene expression and replication in mosquito cells by dsRNA-triggered RNA interference.Mol Ther. 2002 Aug;6(2):243-51. doi: 10.1006/mthe.2002.0652. Mol Ther. 2002. PMID: 12161191
-
Sindbis virus-induced silencing of dengue viruses in mosquitoes.Insect Mol Biol. 2001 Jun;10(3):265-73. doi: 10.1046/j.1365-2583.2001.00267.x. Insect Mol Biol. 2001. PMID: 11437918
-
RNA interference, arthropod-borne viruses, and mosquitoes.Virus Res. 2004 Jun 1;102(1):65-74. doi: 10.1016/j.virusres.2004.01.017. Virus Res. 2004. PMID: 15068882 Review.
-
Molecular strategies for interrupting arthropod-borne virus transmission by mosquitoes.Clin Microbiol Rev. 2000 Oct;13(4):651-61. doi: 10.1128/CMR.13.4.651. Clin Microbiol Rev. 2000. PMID: 11023962 Free PMC article. Review.
Cited by
-
RNA interference for antiviral therapy.J Gene Med. 2006 Aug;8(8):933-50. doi: 10.1002/jgm.929. J Gene Med. 2006. PMID: 16779870 Free PMC article. Review.
-
Short interfering RNA-mediated inhibition of herpes simplex virus type 1 gene expression and function during infection of human keratinocytes.J Virol. 2004 Oct;78(19):10276-81. doi: 10.1128/JVI.78.19.10276-10281.2004. J Virol. 2004. PMID: 15367593 Free PMC article.
-
RNA interference: its use as antiviral therapy.Handb Exp Pharmacol. 2006;173(173):117-50. doi: 10.1007/3-540-27262-3_7. Handb Exp Pharmacol. 2006. PMID: 16594614 Free PMC article. Review.
-
Nucleic acid-based immune system: the antiviral potential of mammalian RNA silencing.J Virol. 2003 Jul;77(13):7159-65. doi: 10.1128/jvi.77.13.7159-7165.2003. J Virol. 2003. PMID: 12805414 Free PMC article. Review. No abstract available.
-
Oral delivery of double-stranded RNA in larvae of the yellow fever mosquito, Aedes aegypti: implications for pest mosquito control.J Insect Sci. 2013;13:69. doi: 10.1673/031.013.6901. J Insect Sci. 2013. PMID: 24224468 Free PMC article.
References
-
- Adelman, Z. N., C. D. Blair, J. O. Carlson, B. J. Beaty, and K. E. Olson. 2001. Sindbis virus-induced silencing of dengue viruses in mosquitoes. Insect Mol. Biol. 10:265-273. - PubMed
-
- Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. - PubMed
-
- Caplen, N. J., Z. Zheng, B. Falgout, and R. A. Morgan. 2002. Inhibition of viral gene expression and replication in mosquito cells by dsRNA-triggered RNA interference. Mol. Ther. 6:243-251. - PubMed
-
- Carlson, J. O., K. E. Olson, S. Higgs, and B. J. Beaty. 1995. Molecular manipulation of mosquitoes. Annu. Rev. Entomol. 40:359-388. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

