Two putative BIN2 substrates are nuclear components of brassinosteroid signaling
- PMID: 12427989
- PMCID: PMC166643
- DOI: 10.1104/pp.102.010918
Two putative BIN2 substrates are nuclear components of brassinosteroid signaling
Abstract
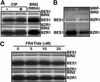
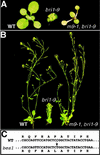
GSK3 is a highly conserved kinase that negatively regulates many cellular processes by phosphorylating a variety of protein substrates. BIN2 is a GSK3-like kinase in Arabidopsis that functions as a negative regulator of brassinosteroid (BR) signaling. It was proposed that BR signals, perceived by a membrane BR receptor complex that contains the leucine (Leu)-rich repeat receptor-like kinase BRI1, inactivate BIN2 to relieve its inhibitory effect on unknown downstream BR-signaling components. Using a yeast (Saccharomyces cerevisiae) two-hybrid approach, we discovered a potential BIN2 substrate that is identical to a recently identified BR-signaling protein, BES1. BES1 and its closest homolog, BZR1, which was also uncovered as a potential BR-signaling protein, display specific interactions with BIN2 in yeast. Both BES1 and BZR1 contain many copies of a conserved GSK3 phosphorylation site and can be phosphorylated by BIN2 in vitro via a novel GSK3 phosphorylation mechanism that is independent of a priming phosphorylation or a scaffold protein. Five independent bes1 alleles containing the same proline-233-Leu mutation were identified as semidominant suppressors of two different bri1 mutations. Over-expression of the wild-type BZR1 gene partially complemented bin2/+ mutants and resulted in a BRI1 overexpression phenotype in a BIN2(+) background, whereas overexpression of a mutated BZR1 gene containing the corresponding proline-234-Leu mutation rescued a weak bri1 mutation and led to a bes1-like phenotype. Confocal microscopic analysis indicated that both BES1 and BZR1 proteins were mainly localized in the nucleus. We propose that BES1/BZR1 are two nuclear components of BR signaling that are negatively regulated by BIN2 through a phosphorylation-initiated process.
Figures




Similar articles
-
Transcription factor HAT1 is phosphorylated by BIN2 kinase and mediates brassinosteroid repressed gene expression in Arabidopsis.Plant J. 2014 Jan;77(1):59-70. doi: 10.1111/tpj.12368. Epub 2013 Dec 3. Plant J. 2014. PMID: 24164091
-
MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis.Proc Natl Acad Sci U S A. 2012 Dec 4;109(49):20142-7. doi: 10.1073/pnas.1205232109. Epub 2012 Nov 19. Proc Natl Acad Sci U S A. 2012. PMID: 23169658 Free PMC article.
-
BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling.Plant Physiol. 2009 Jun;150(2):710-21. doi: 10.1104/pp.109.138099. Epub 2009 Apr 24. Plant Physiol. 2009. PMID: 19395409 Free PMC article.
-
The brassinosteroid signal transduction pathway.Cell Res. 2006 May;16(5):427-34. doi: 10.1038/sj.cr.7310054. Cell Res. 2006. PMID: 16699538 Free PMC article. Review.
-
Regulation of Three Key Kinases of Brassinosteroid Signaling Pathway.Int J Mol Sci. 2020 Jun 18;21(12):4340. doi: 10.3390/ijms21124340. Int J Mol Sci. 2020. PMID: 32570783 Free PMC article. Review.
Cited by
-
Functional characterization of GmBZL2 (AtBZR1 like gene) reveals the conserved BR signaling regulation in Glycine max.Sci Rep. 2016 Aug 8;6:31134. doi: 10.1038/srep31134. Sci Rep. 2016. PMID: 27498784 Free PMC article.
-
Brassinosteroid biosynthesis and signalling in Petunia hybrida.J Exp Bot. 2013 May;64(8):2435-48. doi: 10.1093/jxb/ert102. Epub 2013 Apr 18. J Exp Bot. 2013. PMID: 23599276 Free PMC article.
-
Redundancy, Feedback, and Robustness in the Arabidopsis thaliana BZR/BEH Gene Family.Front Genet. 2018 Nov 13;9:523. doi: 10.3389/fgene.2018.00523. eCollection 2018. Front Genet. 2018. PMID: 30542366 Free PMC article.
-
The RLK/Pelle family of kinases.Plant J. 2011 Apr;66(1):117-27. doi: 10.1111/j.1365-313X.2011.04518.x. Plant J. 2011. PMID: 21443627 Free PMC article. Review.
-
Glycogen synthase kinases in model and crop plants - From negative regulators of brassinosteroid signaling to multifaceted hubs of various signaling pathways and modulators of plant reproduction and yield.Front Plant Sci. 2022 Jul 15;13:939487. doi: 10.3389/fpls.2022.939487. eCollection 2022. Front Plant Sci. 2022. PMID: 35909730 Free PMC article. Review.
References
-
- Clouse SD, Feldmann KA. Molecular genetics of brassinosteroid action. In: Sakurai A, Yokota T, Clouse SD, editors. Brassinosteroids: Steroidal Plant Hormones. Tokyo: Springer-Verlag; 1999. pp. 163–189.
-
- Clouse SD, Sasse JM. Brassinosteroids, essential regulator of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. - PubMed
-
- Cohen P, Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2:769–776. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

