Mapping the encapsidation determinants of feline immunodeficiency virus
- PMID: 12414931
- PMCID: PMC136900
- DOI: 10.1128/jvi.76.23.11889-11903.2002
Mapping the encapsidation determinants of feline immunodeficiency virus
Abstract
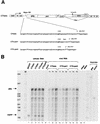
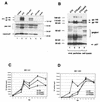
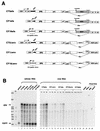
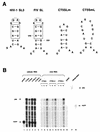
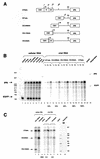
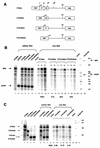
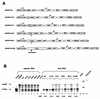
Encapsidation of retroviral RNA involves specific interactions between viral proteins and cis-acting genomic RNA sequences. Human immunodeficiency virus type 1 (HIV-1) RNA encapsidation determinants appear to be more complex and dispersed than those of murine retroviruses. Feline lentiviral (feline immunodeficiency virus [FIV]) encapsidation has not been studied. To gain comparative insight into lentiviral encapsidation and to optimize FIV-based vectors, we used RNase protection assays of cellular and virion RNAs to determine packaging efficiencies of FIV deletion mutants, and we studied replicative phenotypes of mutant viruses. Unlike the case for other mammalian retroviruses, the sequences between the major splice donor (MSD) and the start codon of gag contribute negligibly to FIV encapsidation. Moreover, molecular clones having deletions in this region were replication competent. In contrast, sequences upstream of the MSD were important for encapsidation, and deletion of the U5 element markedly reduced genomic RNA packaging. The contribution of gag sequences to packaging was systematically investigated with subgenomic FIV vectors containing variable portions of the gag open reading frame, with all virion proteins supplied in trans. When no gag sequence was present, packaging was abolished and marker gene transduction was absent. Inclusion of the first 144 nucleotides (nt) of gag increased vector encapsidation to detectable levels, while inclusion of the first 311 nt increased it to nearly wild-type levels and resulted in high-titer FIV vectors. However, the identified proximal gag sequence is necessary but not sufficient, since viral mRNAs that contain all coding regions, with or without as much as 119 nt of adjacent upstream 5' leader, were excluded from encapsidation. The results identify a mechanism whereby FIV can encapsidate its genomic mRNA in preference to subgenomic mRNAs.
Figures


Similar articles
-
The critical role of proximal gag sequences in feline immunodeficiency virus genome encapsidation.Virology. 2004 Sep 15;327(1):111-20. doi: 10.1016/j.virol.2004.06.014. Virology. 2004. PMID: 15327902
-
Human immunodeficiency virus type 2 (HIV-2): packaging signal and associated negative regulatory element.Hum Gene Ther. 1995 Feb;6(2):177-84. doi: 10.1089/hum.1995.6.2-177. Hum Gene Ther. 1995. PMID: 7734518
-
Sequences within the gag gene of feline immunodeficiency virus (FIV) are important for efficient RNA encapsidation.Virus Res. 2003 Jun;93(2):199-209. doi: 10.1016/s0168-1702(03)00098-4. Virus Res. 2003. PMID: 12782368
-
Properties and Functions of Feline Immunodeficiency Virus Gag Domains in Virion Assembly and Budding.Viruses. 2018 May 16;10(5):261. doi: 10.3390/v10050261. Viruses. 2018. PMID: 29772651 Free PMC article. Review.
-
FIV Gag: virus assembly and host-cell interactions.Vet Immunol Immunopathol. 2010 Mar 15;134(1-2):3-13. doi: 10.1016/j.vetimm.2009.10.003. Epub 2009 Oct 14. Vet Immunol Immunopathol. 2010. PMID: 19910057 Free PMC article. Review.
Cited by
-
Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species.Proc Natl Acad Sci U S A. 2008 Mar 4;105(9):3563-8. doi: 10.1073/pnas.0709258105. Epub 2008 Feb 19. Proc Natl Acad Sci U S A. 2008. PMID: 18287034 Free PMC article.
-
The molecular biology of feline immunodeficiency virus (FIV).Viruses. 2011 Nov;3(11):2192-213. doi: 10.3390/v3112192. Epub 2011 Nov 9. Viruses. 2011. PMID: 22163340 Free PMC article. Review.
-
Streamlined design of a self-inactivating feline immunodeficiency virus vector for transducing ex vivo dendritic cells and T lymphocytes.Genet Vaccines Ther. 2007 Sep 19;5:8. doi: 10.1186/1479-0556-5-8. Genet Vaccines Ther. 2007. PMID: 17880683 Free PMC article.
-
Optimal packaging of FIV genomic RNA depends upon a conserved long-range interaction and a palindromic sequence within gag.J Mol Biol. 2010 Oct 15;403(1):103-119. doi: 10.1016/j.jmb.2010.08.019. Epub 2010 Aug 21. J Mol Biol. 2010. PMID: 20732330 Free PMC article.
-
Feline immunodeficiency virus Gag is a nuclear shuttling protein.J Virol. 2012 Aug;86(16):8402-11. doi: 10.1128/JVI.00692-12. Epub 2012 May 23. J Virol. 2012. PMID: 22623802 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

