The p127 subunit (DDB1) of the UV-DNA damage repair binding protein is essential for the targeted degradation of STAT1 by the V protein of the paramyxovirus simian virus 5
- PMID: 12388698
- PMCID: PMC136798
- DOI: 10.1128/jvi.76.22.11379-11386.2002
The p127 subunit (DDB1) of the UV-DNA damage repair binding protein is essential for the targeted degradation of STAT1 by the V protein of the paramyxovirus simian virus 5
Abstract
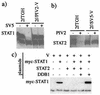
The V protein of simian virus 5 (SV5) blocks interferon signaling by targeting STAT1 for proteasome-mediated degradation. Here we present three main pieces of evidence which demonstrate that the p127 subunit (DDB1) of the UV damage-specific DNA binding protein (DDB) plays a central role in this degradation process. First, the V protein of an SV5 mutant which fails to target STAT1 for degradation does not bind DDB1. Second, mutations in the N and C termini of V which abolish the binding of V to DDB1 also prevent V from blocking interferon (IFN) signaling. Third, treatment of HeLa/SV5-V cells, which constitutively express the V protein of SV5 and thus lack STAT1, with short interfering RNAs specific for DDB1 resulted in a reduction in DDB1 levels with a concomitant increase in STAT1 levels and a restoration of IFN signaling. Furthermore, STAT1 is degraded in GM02415 (2RO) cells, which have a mutation in DDB2 (the p48 subunit of DDB) which abolishes its ability to interact with DDB1, thereby demonstrating that the role of DDB1 in STAT1 degradation is independent of its association with DDB2. Evidence is also presented which demonstrates that STAT2 is required for the degradation of STAT1 by SV5. These results suggest that DDB1, STAT1, STAT2, and V may form part of a large multiprotein complex which leads to the targeted degradation of STAT1 by the proteasome.
Figures
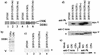



Similar articles
-
Hepatitis B virus X protein and simian virus 5 V protein exhibit similar UV-DDB1 binding properties to mediate distinct activities.J Virol. 2003 Jun;77(11):6274-83. doi: 10.1128/jvi.77.11.6274-6283.2003. J Virol. 2003. PMID: 12743284 Free PMC article.
-
Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons.J Virol. 2002 Mar;76(5):2159-67. doi: 10.1128/jvi.76.5.2159-2167.2002. J Virol. 2002. PMID: 11836393 Free PMC article.
-
Simian virus 5 V protein acts as an adaptor, linking DDB1 to STAT2, to facilitate the ubiquitination of STAT1.J Virol. 2005 Nov;79(21):13434-41. doi: 10.1128/JVI.79.21.13434-13441.2005. J Virol. 2005. PMID: 16227264 Free PMC article.
-
DDB complexities.DNA Repair (Amst). 2003 Sep 18;2(9):1065-9. doi: 10.1016/s1568-7864(03)00113-7. DNA Repair (Amst). 2003. PMID: 12967661 Review.
-
Paramyxovirus strategies for evading the interferon response.Rev Med Virol. 2002 Nov-Dec;12(6):337-57. doi: 10.1002/rmv.357. Rev Med Virol. 2002. PMID: 12410527 Review.
Cited by
-
Paramyxovirus activation and inhibition of innate immune responses.J Mol Biol. 2013 Dec 13;425(24):4872-92. doi: 10.1016/j.jmb.2013.09.015. Epub 2013 Sep 20. J Mol Biol. 2013. PMID: 24056173 Free PMC article. Review.
-
Human parainfluenza virus type 2 V protein inhibits genome replication by binding to the L protein: possible role in promoting viral fitness.J Virol. 2008 Jul;82(13):6130-8. doi: 10.1128/JVI.02635-07. Epub 2008 Apr 16. J Virol. 2008. PMID: 18417591 Free PMC article.
-
Molecular insights into NF2/Merlin tumor suppressor function.FEBS Lett. 2014 Aug 19;588(16):2743-52. doi: 10.1016/j.febslet.2014.04.001. Epub 2014 Apr 12. FEBS Lett. 2014. PMID: 24726726 Free PMC article. Review.
-
Evasion of Host Antiviral Innate Immunity by Paramyxovirus Accessory Proteins.Front Microbiol. 2022 Jan 31;12:790191. doi: 10.3389/fmicb.2021.790191. eCollection 2021. Front Microbiol. 2022. PMID: 35173691 Free PMC article. Review.
-
HIV-1 Vpr-mediated G2 arrest involves the DDB1-CUL4AVPRBP E3 ubiquitin ligase.PLoS Pathog. 2007 Jul;3(7):e85. doi: 10.1371/journal.ppat.0030085. PLoS Pathog. 2007. PMID: 17630831 Free PMC article.
References
-
- Andrejeva, J., D. F. Young, S. Goodbourn, and R. E. Randall. 2002. Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. J. Virol. 76:2159-2167. - PMC - PubMed
-
- Bartel, P. L., C.-T. Chien, R. Sternglanz and S. Fields. 1993. Using the two-hybrid system to detect protein-protein interactions, p. 153-179. Cellular interactions in development: a practical approach. Oxford University Press, Oxford, United Kingdom.
-
- Chatziandreou, N., D. Young, J. Andrejeva, S. Goodbourn, and R. E. Randall. 2002. Differences in interferon sensitivity and biological properties of two related isolates of simian virus 5: a model for virus persistence. Virology 293:234-242. - PubMed
-
- Chen, X., Y. Zhang, L. Douglas, and P. Zhou. 2001. UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J. Biol. Chem. 276:48175-48182. - PubMed
-
- Choppin, P. W. 1964. Multiplication of a myxovirus (SV5) with minimal cytopathic effects and without interference. Virology 23:224-233. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

