The osteopetrotic mutation toothless (tl) is a loss-of-function frameshift mutation in the rat Csf1 gene: Evidence of a crucial role for CSF-1 in osteoclastogenesis and endochondral ossification
- PMID: 12379742
- PMCID: PMC137879
- DOI: 10.1073/pnas.202332999
The osteopetrotic mutation toothless (tl) is a loss-of-function frameshift mutation in the rat Csf1 gene: Evidence of a crucial role for CSF-1 in osteoclastogenesis and endochondral ossification
Abstract
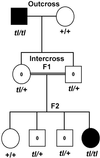
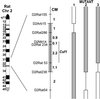
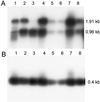
The toothless (tl) mutation in the rat is a naturally occurring, autosomal recessive mutation resulting in a profound deficiency of bone-resorbing osteoclasts and peritoneal macrophages. The failure to resorb bone produces severe, unrelenting osteopetrosis, with a highly sclerotic skeleton, lack of marrow spaces, failure of tooth eruption, and other pathologies. Injections of CSF-1 improve some, but not all, of these. In this report we have used polymorphism mapping, sequencing, and expression studies to identify the genetic lesion in the tl rat. We found a 10-base insertion near the beginning of the open reading of the Csf1 gene that yields a truncated, nonfunctional protein and an early stop codon, thus rendering the tl rat CSF-1(null). All mutants were homozygous for the mutation and all carriers were heterozygous. No CSF-1 transcripts were identified in rat mRNA that would avoid the mutation via alternative splicing. The biology and actions of CSF-1 have been elucidated by many studies that use another naturally occurring mutation, the op mouse, in which a single base insertion also disrupts the reading frame. The op mouse has milder osteoclastopenia and osteopetrosis than the tl rat and recovers spontaneously over the first few months of life. Thus, the tl rat provides a second model in which the functions of CSF-1 can be studied. Understanding the similarities and differences in the phenotypes of these two models will be important to advancing our knowledge of the many actions of CSF-1.
Figures





Similar articles
-
Mutation of macrophage colony stimulating factor (Csf1) causes osteopetrosis in the tl rat.Biochem Biophys Res Commun. 2002 Jun 28;294(5):1114-20. doi: 10.1016/S0006-291X(02)00598-3. Biochem Biophys Res Commun. 2002. PMID: 12074592
-
Heterogeneity of colony stimulating factor-1 gene expression in the skeleton of four osteopetrotic mutations in rats and mice.J Cell Physiol. 1996 Feb;166(2):340-50. doi: 10.1002/(SICI)1097-4652(199602)166:2<340::AID-JCP12>3.0.CO;2-F. J Cell Physiol. 1996. PMID: 8591994
-
A new histomorphometric method to assess growth plate chondrodysplasia and its application to the toothless (tl, Csf1(null)) osteopetrotic rat.Connect Tissue Res. 2004;45(1):1-10. doi: 10.1080/03008200490278016. Connect Tissue Res. 2004. PMID: 15203935
-
Recent developments in the understanding of the pathophysiology of osteopetrosis.Eur J Endocrinol. 1996 Feb;134(2):143-56. doi: 10.1530/eje.0.1340143. Eur J Endocrinol. 1996. PMID: 8630510 Review.
-
Role of CSF-1 in bone and bone marrow development.Mol Reprod Dev. 1997 Jan;46(1):75-83; discussion 83-4. doi: 10.1002/(SICI)1098-2795(199701)46:1<75::AID-MRD12>3.0.CO;2-2. Mol Reprod Dev. 1997. PMID: 8981367 Review.
Cited by
-
Unique and non-redundant function of csf1r paralogues in regulation and evolution of post-embryonic development of the zebrafish.Development. 2020 Jan 22;147(2):dev181834. doi: 10.1242/dev.181834. Development. 2020. PMID: 31932352 Free PMC article.
-
Sost Deficiency does not Alter Bone's Lacunar or Vascular Porosity in Mice.Front Mater. 2017 Sep 13;4:27. doi: 10.3389/fmats.2017.00027. Front Mater. 2017. PMID: 29349060 Free PMC article.
-
Phospho1 deficiency transiently modifies bone architecture yet produces consistent modification in osteocyte differentiation and vascular porosity with ageing.Bone. 2015 Dec;81:277-291. doi: 10.1016/j.bone.2015.07.035. Epub 2015 Jul 29. Bone. 2015. PMID: 26232374 Free PMC article.
-
Deciphering the roles of macrophages in developmental and inflammation stimulated lymphangiogenesis.Vasc Cell. 2012 Sep 3;4(1):15. doi: 10.1186/2045-824X-4-15. Vasc Cell. 2012. PMID: 22943568 Free PMC article.
-
M-CSFR/CSF1R signaling regulates myeloid fates in zebrafish via distinct action of its receptors and ligands.Blood Adv. 2022 Mar 8;6(5):1474-1488. doi: 10.1182/bloodadvances.2021005459. Blood Adv. 2022. PMID: 34979548 Free PMC article.
References
-
- Marks S. C. & Odgren, P. R. (2002) in Principles of Bone Biology, eds. Bilezikian, J. P., Raisz, L. G. & Rodan, G. A. (Academic, New York), Vol. 1, pp. 3–15.
-
- Popoff S. N. & Schneider, G. B. (1996) Mol. Med. Today 2, 349-358. - PubMed
-
- Stanley E. R., Chen, D. M. & Lin, H. S. (1978) Nature 274, 168-170. - PubMed
-
- Yoshida H., Hayashi, S., Kunisada, T., Ogawa, M., Nishikawa, S., Okamura, H., Sudo, T., Shultz, L. D. & Nishikawa, S. (1990) Nature 345, 442-444. - PubMed
-
- Wong B. R., Rho, J., Arron, J., Robinson, E., Orlinick, J., Chao, M., Kalachikov, S., Cayani, E., Bartlett, F. S., III, Frankel, W. N., et al. (1997) J. Biol. Chem. 272, 25190-25194. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

