RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach
- PMID: 12242279
- PMCID: PMC139818
- DOI: 10.1128/MCB.22.20.6979-6992.2002
RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach
Abstract
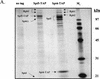
To physically characterize the web of interactions connecting the Saccharomyces cerevisiae proteins suspected to be RNA polymerase II (RNAPII) elongation factors, subunits of Spt4/Spt5 and Spt16/Pob3 (corresponding to human DSIF and FACT), Spt6, TFIIF (Tfg1, -2, and -3), TFIIS, Rtf1, and Elongator (Elp1, -2, -3, -4, -5, and -6) were affinity purified under conditions designed to minimize loss of associated polypeptides and then identified by mass spectrometry. Spt16/Pob3 was discovered to associate with three distinct complexes: histones; Chd1/casein kinase II (CKII); and Rtf1, Paf1, Ctr9, Cdc73, and a previously uncharacterized protein, Leo1. Rtf1 and Chd1 have previously been implicated in the control of elongation, and the sensitivity to 6-azauracil of strains lacking Paf1, Cdc73, or Leo1 suggested that these proteins are involved in elongation by RNAPII as well. Confirmation came from chromatin immunoprecipitation (ChIP) assays demonstrating that all components of this complex, including Leo1, cross-linked to the promoter, coding region, and 3' end of the ADH1 gene. In contrast, the three subunits of TFIIF cross-linked only to the promoter-containing fragment of ADH1. Spt6 interacted with the uncharacterized, essential protein Iws1 (interacts with Spt6), and Spt5 interacted either with Spt4 or with a truncated form of Spt6. ChIP on Spt6 and the novel protein Iws1 resulted in the cross-linking of both proteins to all three regions of the ADH1 gene, suggesting that Iws1 is likely an Spt6-interacting elongation factor. Spt5, Spt6, and Iws1 are phosphorylated on consensus CKII sites in vivo, conceivably by the Chd1/CKII associated with Spt16/Pob3. All the elongation factors but Elongator copurified with RNAPII.
Figures


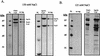
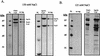
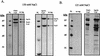
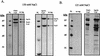
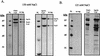


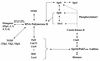
Similar articles
-
The Paf1 complex physically and functionally associates with transcription elongation factors in vivo.EMBO J. 2002 Apr 2;21(7):1764-74. doi: 10.1093/emboj/21.7.1764. EMBO J. 2002. PMID: 11927560 Free PMC article.
-
Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae.Genes Dev. 1998 Feb 1;12(3):357-69. doi: 10.1101/gad.12.3.357. Genes Dev. 1998. PMID: 9450930 Free PMC article.
-
Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins.Mol Cell Biol. 2003 Feb;23(4):1368-78. doi: 10.1128/MCB.23.4.1368-1378.2003. Mol Cell Biol. 2003. PMID: 12556496 Free PMC article.
-
The Paf1 complex: platform or player in RNA polymerase II transcription?Biochim Biophys Acta. 2010 May-Jun;1799(5-6):379-88. doi: 10.1016/j.bbagrm.2010.01.001. Epub 2010 Jan 12. Biochim Biophys Acta. 2010. PMID: 20060942 Free PMC article. Review.
-
Mechanisms of Transcription Elongation Factor DSIF (Spt4-Spt5).J Mol Biol. 2021 Jul 9;433(14):166657. doi: 10.1016/j.jmb.2020.09.016. Epub 2020 Sep 25. J Mol Biol. 2021. PMID: 32987031 Review.
Cited by
-
Functional analysis of Bre1p, an E3 ligase for histone H2B ubiquitylation, in regulation of RNA polymerase II association with active genes and transcription in vivo.J Biol Chem. 2013 Apr 5;288(14):9619-9633. doi: 10.1074/jbc.M113.450403. Epub 2013 Feb 15. J Biol Chem. 2013. PMID: 23417674 Free PMC article.
-
Structural basis of nucleosome transcription mediated by Chd1 and FACT.Nat Struct Mol Biol. 2021 Apr;28(4):382-387. doi: 10.1038/s41594-021-00578-6. Epub 2021 Apr 12. Nat Struct Mol Biol. 2021. PMID: 33846633 Free PMC article.
-
The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states.Biochim Biophys Acta. 2013 Jan;1829(1):116-26. doi: 10.1016/j.bbagrm.2012.08.011. Epub 2012 Sep 6. Biochim Biophys Acta. 2013. PMID: 22982193 Free PMC article.
-
Histone Chaperones Spt6 and FACT: Similarities and Differences in Modes of Action at Transcribed Genes.Genet Res Int. 2011;2011:625210. doi: 10.4061/2011/625210. Epub 2011 Oct 26. Genet Res Int. 2011. PMID: 22567361 Free PMC article.
-
Chromatin and transcription in yeast.Genetics. 2012 Feb;190(2):351-87. doi: 10.1534/genetics.111.132266. Genetics. 2012. PMID: 22345607 Free PMC article. Review.
References
-
- Bähler, J., J. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
