A P22 scaffold protein mutation increases the robustness of head assembly in the presence of excess portal protein
- PMID: 12239300
- PMCID: PMC136566
- DOI: 10.1128/jvi.76.20.10245-10255.2002
A P22 scaffold protein mutation increases the robustness of head assembly in the presence of excess portal protein
Abstract
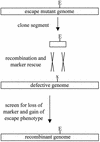
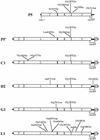
Bacteriophage with linear, double-stranded DNA genomes package DNA into preassembled protein shells called procapsids. Located at one vertex in the procapsid is a portal complex composed of a ring of 12 subunits of portal protein. The portal complex serves as a docking site for the DNA packaging enzymes, a conduit for the passage of DNA, and a binding site for the phage tail. An excess of the P22 portal protein alters the assembly pathway of the procapsid, giving rise to defective procapsid-like particles and aberrant heads. In the present study, we report the isolation of escape mutant phage that are able to replicate more efficiently than wild-type phage in the presence of excess portal protein. The escape mutations all mapped to the same phage genome segment spanning the portal, scaffold, coat, and open reading frame 69 genes. The mutations present in five of the escape mutants were determined by DNA sequencing. Interestingly, each mutant contained the same mutation in the scaffold gene, which changes the glycine at position 287 to glutamate. This mutation alone conferred an escape phenotype, and the heads assembled by phage harboring only this mutation had reduced levels of portal protein and exhibited increased head assembly fidelity in the presence of excess portal protein. Because this mutation resides in a region of scaffold protein necessary for coat protein binding, these findings suggest that the P22 scaffold protein may define the portal vertices in an indirect manner, possibly by regulating the fidelity of coat protein polymerization.
Figures




Similar articles
-
Bacteriophage p22 portal vertex formation in vivo.J Mol Biol. 2002 Feb 1;315(5):975-94. doi: 10.1006/jmbi.2001.5275. J Mol Biol. 2002. PMID: 11827470
-
Molecular genetics of bacteriophage P22 scaffolding protein's functional domains.J Mol Biol. 2005 May 13;348(4):831-44. doi: 10.1016/j.jmb.2005.03.004. J Mol Biol. 2005. PMID: 15843016
-
A Molecular Staple: D-Loops in the I Domain of Bacteriophage P22 Coat Protein Make Important Intercapsomer Contacts Required for Procapsid Assembly.J Virol. 2015 Oct;89(20):10569-79. doi: 10.1128/JVI.01629-15. Epub 2015 Aug 12. J Virol. 2015. PMID: 26269173 Free PMC article.
-
'Let the phage do the work': using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants.Virology. 2010 Jun 5;401(2):119-30. doi: 10.1016/j.virol.2010.02.017. Epub 2010 Mar 16. Virology. 2010. PMID: 20236676 Free PMC article. Review.
-
Phage assembly and the special role of the portal protein.Curr Opin Virol. 2018 Aug;31:66-73. doi: 10.1016/j.coviro.2018.09.004. Epub 2018 Sep 28. Curr Opin Virol. 2018. PMID: 30274853 Review.
Cited by
-
Portal Protein: The Orchestrator of Capsid Assembly for the dsDNA Tailed Bacteriophages and Herpesviruses.Annu Rev Virol. 2019 Sep 29;6(1):141-160. doi: 10.1146/annurev-virology-092818-015819. Epub 2019 Jul 23. Annu Rev Virol. 2019. PMID: 31337287 Free PMC article. Review.
-
Functional metagenomics of the thioredoxin superfamily.J Biol Chem. 2021 Jan-Jun;296:100247. doi: 10.1074/jbc.RA120.016350. Epub 2021 Jan 14. J Biol Chem. 2021. PMID: 33361108 Free PMC article.
-
Involvement of the portal at an early step in herpes simplex virus capsid assembly.J Virol. 2005 Aug;79(16):10540-6. doi: 10.1128/JVI.79.16.10540-10546.2005. J Virol. 2005. PMID: 16051846 Free PMC article.
-
Clostridioides difficile phage biology and application.FEMS Microbiol Rev. 2021 Sep 8;45(5):fuab012. doi: 10.1093/femsre/fuab012. FEMS Microbiol Rev. 2021. PMID: 33580957 Free PMC article. Review.
-
Tryptophan Residues Are Critical for Portal Protein Assembly and Incorporation in Bacteriophage P22.Viruses. 2022 Jun 27;14(7):1400. doi: 10.3390/v14071400. Viruses. 2022. PMID: 35891382 Free PMC article.
References
-
- Bazinet, C., and J. King. 1985. The DNA translocating vertex of dsDNA bacteriophage. Annu. Rev. Microbiol. 39:109-129. - PubMed
-
- Bazinet, C., and J. King. 1988. Initiation of P22 procapsid assembly in vivo. J. Mol. Biol. 202:77-86. - PubMed
-
- Bazinet, C., R. Villafane, and J. King. 1990. Novel second-site suppression of a cold-sensitive defect in phage P22 procapsid assembly. J. Mol. Biol. 216:701-716. - PubMed
-
- Botstein, D., and M. Levine. 1968. Intermediates in the synthesis of phage P22 DNA. Cold Spring Harbor Symp. Quant. Biol. 33:659-667. - PubMed
-
- Botstein, D., C. H. Waddell, and J. King. 1973. Mechanism of head assembly and DNA encapsulation in Salmonella phage p22. I. Genes, proteins, structures and DNA maturation. J. Mol. Biol. 80:669-695. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

