Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by the viral protease
- PMID: 12239298
- PMCID: PMC136535
- DOI: 10.1128/jvi.76.20.10226-10233.2002
Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by the viral protease
Abstract
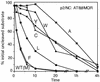
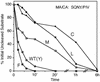
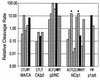
Processing of the human immunodeficiency virus type 1 (HIV-1) Gag precursor is highly regulated, with differential rates of cleavage at the five major processing sites to give characteristic processing intermediates. We examined the role of the P1 amino acid in determining the rate of cleavage at each of these five sites by using libraries of mutants generated by site-directed mutagenesis. Between 12 and 17 substitution mutants were tested at each P1 position in Gag, using recombinant HIV-1 protease (PR) in an in vitro processing reaction of radiolabeled Gag substrate. There were three sites in Gag (MA/CA, CA/p2, NC/p1) where one or more substitutions mediated enhanced rates of cleavage, with an enhancement greater than 60-fold in the case of NC/p1. For the other two sites (p2/NC, p1/p6), the wild-type amino acid conferred optimal cleavage. The order of the relative rates of cleavage with the P1 amino acids Tyr, Met, and Leu suggests that processing sites can be placed into two groups and that the two groups are defined by the size of the P1' amino acid. These results point to a trans effect between the P1 and P1' amino acids that is likely to be a major determinant of the rate of cleavage at the individual sites and therefore also a determinant of the ordered cleavage of the Gag precursor.
Figures


Similar articles
-
Context surrounding processing sites is crucial in determining cleavage rate of a subset of processing sites in HIV-1 Gag and Gag-Pro-Pol polyprotein precursors by viral protease.J Biol Chem. 2012 Apr 13;287(16):13279-90. doi: 10.1074/jbc.M112.339374. Epub 2012 Feb 13. J Biol Chem. 2012. PMID: 22334652 Free PMC article.
-
Naturally occurring amino acid polymorphisms in human immunodeficiency virus type 1 (HIV-1) Gag p7(NC) and the C-cleavage site impact Gag-Pol processing by HIV-1 protease.Virology. 2002 Jan 5;292(1):137-49. doi: 10.1006/viro.2001.1184. Virology. 2002. PMID: 11878916
-
Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites.J Virol. 1997 Sep;71(9):6662-70. doi: 10.1128/JVI.71.9.6662-6670.1997. J Virol. 1997. PMID: 9261388 Free PMC article.
-
Processing of retroviral Gag polyproteins: an in vitro approach.Methods Enzymol. 1994;241:227-53. doi: 10.1016/0076-6879(94)41067-4. Methods Enzymol. 1994. PMID: 7854180 Review. No abstract available.
-
Activation mechanism of pepsinogen as compared to the processing of HIV protease gag-pol precursor protein.Adv Exp Med Biol. 1998;436:245-52. doi: 10.1007/978-1-4615-5373-1_34. Adv Exp Med Biol. 1998. PMID: 9561226 Review. No abstract available.
Cited by
-
HIV-1 Maturation: Lessons Learned from Inhibitors.Viruses. 2020 Aug 26;12(9):940. doi: 10.3390/v12090940. Viruses. 2020. PMID: 32858867 Free PMC article. Review.
-
Role of invariant Thr80 in human immunodeficiency virus type 1 protease structure, function, and viral infectivity.J Virol. 2006 Jul;80(14):6906-16. doi: 10.1128/JVI.01900-05. J Virol. 2006. PMID: 16809296 Free PMC article.
-
A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism.PLoS Med. 2007 Jan;4(1):e36. doi: 10.1371/journal.pmed.0040036. PLoS Med. 2007. PMID: 17227139 Free PMC article.
-
Ordered processing of the human immunodeficiency virus type 1 GagPol precursor is influenced by the context of the embedded viral protease.J Virol. 2005 Aug;79(16):10601-7. doi: 10.1128/JVI.79.16.10601-10607.2005. J Virol. 2005. PMID: 16051852 Free PMC article.
-
Processing sites in the human immunodeficiency virus type 1 (HIV-1) Gag-Pro-Pol precursor are cleaved by the viral protease at different rates.Retrovirology. 2005 Nov 1;2:66. doi: 10.1186/1742-4690-2-66. Retrovirology. 2005. PMID: 16262906 Free PMC article.
References
-
- Billich, A., and G. Winkler. 1991. Analysis of subsite preferences of HIV-1 proteinase using MA/CA junction peptides substituted at the P3-P1′ positions. Arch. Biochem. Biophys. 290:186-190. - PubMed
-
- Billich, S., M. T. Knoop, J. Hansen, P. Strop, J. Sedlacek, R. Mertz, and K. Moelling. 1988. Synthetic peptides as substrates and inhibitors of human immune deficiency virus-1 protease. J. Biol. Chem. 263:17905-17908. - PubMed
-
- Cameron, C. E., B. Grinde, P. Jacques, J. Jentoft, J. Leis, A. Wlodawer, and I. T. Weber. 1993. Comparison of the substrate-binding pockets of the Rous sarcoma virus and human immunodeficiency virus type 1 proteases. J. Biol. Chem. 268:11711-11720. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

