Characterisation of site-biased DNA methyltransferases: specificity, affinity and subsite relationships
- PMID: 12202767
- PMCID: PMC137423
- DOI: 10.1093/nar/gkf501
Characterisation of site-biased DNA methyltransferases: specificity, affinity and subsite relationships
Abstract
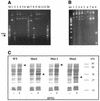
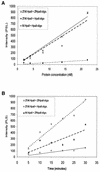
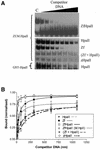
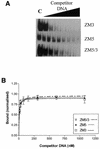
DNA methylation is now seen as a primary signal in the cell for mediating transcriptional repression through chromatin formation. The construction and evaluation of enzymes capable of influencing this process in vivo is therefore of significant interest. We have fused the C5-cytosine DNA methyltransferases, M.HhaI and M.HpaII, which both methylate 4 bp sequences containing a CpG dinucleotide, to a three zinc finger protein recognising a 9 bp DNA sequence. DNA methylation analyses demonstrate specific DNA methylation by both enzymes at target sites comprising adjacent methyltransferase and zinc finger subsites, targeted M.HpaII being the most specific. Binding analysis of the targeted M.HpaII enzyme reveals an 8-fold preference for binding to its target site, compared to binding to a zinc finger site alone, and an 18-fold preference over binding to a methyltransferase site alone, thereby demonstrating enhanced binding by the fusion protein, compared to its component proteins. Both DNA binding and methylation are specific for the target site up to separations of approximately 40 bp between the zinc finger and methyltransferase subsites. Ex vivo plasmid methylation experiments are also described that demonstrate targeted methylation. These targeted enzymes, however, are shown to be not fully mono-functional, retaining a significant non-targeted activity most evident at elevated protein concentrations.
Figures


Similar articles
-
Site-selective in vivo targeting of cytosine-5 DNA methylation by zinc-finger proteins.Nucleic Acids Res. 2003 Nov 15;31(22):6493-501. doi: 10.1093/nar/gkg853. Nucleic Acids Res. 2003. PMID: 14602907 Free PMC article.
-
Specific targeting of cytosine methylation to DNA sequences in vivo.Nucleic Acids Res. 2007;35(3):740-54. doi: 10.1093/nar/gkl1053. Epub 2006 Dec 20. Nucleic Acids Res. 2007. PMID: 17182629 Free PMC article.
-
In vitro DNA cytosine methylation of cis-regulatory elements modulates c-Ha-ras promoter activity in vivo.Nucleic Acids Res. 1989 Jul 11;17(13):5135-47. doi: 10.1093/nar/17.13.5135. Nucleic Acids Res. 1989. PMID: 2474794 Free PMC article.
-
Structure, function, and mechanism of HhaI DNA methyltransferases.Crit Rev Biochem Mol Biol. 2002;37(3):167-97. doi: 10.1080/10409230290771492. Crit Rev Biochem Mol Biol. 2002. PMID: 12139442 Review.
-
Structure and function of DNA methyltransferases.Annu Rev Biophys Biomol Struct. 1995;24:293-318. doi: 10.1146/annurev.bb.24.060195.001453. Annu Rev Biophys Biomol Struct. 1995. PMID: 7663118 Review.
Cited by
-
Site-selective in vivo targeting of cytosine-5 DNA methylation by zinc-finger proteins.Nucleic Acids Res. 2003 Nov 15;31(22):6493-501. doi: 10.1093/nar/gkg853. Nucleic Acids Res. 2003. PMID: 14602907 Free PMC article.
-
Genomic and epigenomic instability, fragile sites, schizophrenia and autism.Curr Genomics. 2010 Sep;11(6):447-69. doi: 10.2174/138920210793176001. Curr Genomics. 2010. PMID: 21358990 Free PMC article.
-
Targeted DNA methylation in human cells using engineered dCas9-methyltransferases.Sci Rep. 2017 Jul 27;7(1):6732. doi: 10.1038/s41598-017-06757-0. Sci Rep. 2017. PMID: 28751638 Free PMC article.
-
Specific targeting of cytosine methylation to DNA sequences in vivo.Nucleic Acids Res. 2007;35(3):740-54. doi: 10.1093/nar/gkl1053. Epub 2006 Dec 20. Nucleic Acids Res. 2007. PMID: 17182629 Free PMC article.
-
Lowering DNA binding affinity of SssI DNA methyltransferase does not enhance the specificity of targeted DNA methylation in E. coli.Sci Rep. 2021 Jul 27;11(1):15226. doi: 10.1038/s41598-021-94528-3. Sci Rep. 2021. PMID: 34315949 Free PMC article.
References
-
- Wilson G. (1992) Amino acid sequence arrangements of DNA-methyltransferases. Methods Enzymol., 216, 259–278. - PubMed
-
- Klimasauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. - PubMed
-
- Reinisch K.M., Chen,L., Verdine,G.L. and Lipscomb,W.N. (1995) The crystal structure of HaeIII methyltransferase covalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell, 82, 143–153. - PubMed
-
- Xu G.-L. and Bestor,T.H. (1997) Cytosine methylation targetted to predetermined sequences. Nature Genet., 17, 376–378. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

