Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription
- PMID: 12195017
- PMCID: PMC129314
- DOI: 10.1073/pnas.182256799
Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription
Abstract
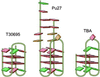
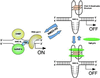
The nuclease hypersensitivity element III(1) upstream of the P1 promoter of c-MYC controls 85-90% of the transcriptional activation of this gene. We have demonstrated that the purine-rich strand of the DNA in this region can form two different intramolecular G-quadruplex structures, only one of which seems to be biologically relevant. This biologically relevant structure is the kinetically favored chair-form G-quadruplex, which is destabilized when mutated with a single G --> A transition, resulting in a 3-fold increase in basal transcriptional activity of the c-MYC promoter. The cationic porphyrin TMPyP4, which has been shown to stabilize this G-quadruplex structure, is able to suppress further c-MYC transcriptional activation. These results provide compelling evidence that a specific G-quadruplex structure formed in the c-MYC promoter region functions as a transcriptional repressor element. Furthermore, we establish the principle that c-MYC transcription can be controlled by ligand-mediated G-quadruplex stabilization.
Figures




Similar articles
-
Drug targeting of the c-MYC promoter to repress gene expression via a G-quadruplex silencer element.Semin Oncol. 2006 Aug;33(4):498-512. doi: 10.1053/j.seminoncol.2006.04.012. Semin Oncol. 2006. PMID: 16890804 Review.
-
The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4.J Am Chem Soc. 2004 Jul 21;126(28):8702-9. doi: 10.1021/ja040022b. J Am Chem Soc. 2004. PMID: 15250722
-
Stabilization of the c-myc gene promoter quadruplex by specific ligands' inhibitors of telomerase.Biochem Biophys Res Commun. 2004 Oct 22;323(3):802-8. doi: 10.1016/j.bbrc.2004.08.150. Biochem Biophys Res Commun. 2004. PMID: 15381071
-
Effects of triethylene tetraamine on the G-quadruplex structure in the human c-myc promoter.J Biochem. 2007 May;141(5):669-74. doi: 10.1093/jb/mvm069. Epub 2007 Mar 4. J Biochem. 2007. PMID: 17339229
-
Structure of the biologically relevant G-quadruplex in the c-MYC promoter.Nucleosides Nucleotides Nucleic Acids. 2006;25(8):951-68. doi: 10.1080/15257770600809913. Nucleosides Nucleotides Nucleic Acids. 2006. PMID: 16901825 Review.
Cited by
-
Genome-wide discovery of G-quadruplexes in barley.Sci Rep. 2021 Apr 12;11(1):7876. doi: 10.1038/s41598-021-86838-3. Sci Rep. 2021. PMID: 33846409 Free PMC article.
-
Beyond small molecules: targeting G-quadruplex structures with oligonucleotides and their analogues.Nucleic Acids Res. 2021 Jul 9;49(12):6638-6659. doi: 10.1093/nar/gkab334. Nucleic Acids Res. 2021. PMID: 33978760 Free PMC article. Review.
-
CX-5461 Preferentially Induces Top2α-Dependent DNA Breaks at Ribosomal DNA Loci.Biomedicines. 2024 Jul 8;12(7):1514. doi: 10.3390/biomedicines12071514. Biomedicines. 2024. PMID: 39062087 Free PMC article.
-
The G4 Resolvase DHX36 Possesses a Prognosis Significance and Exerts Tumour Suppressing Function Through Multiple Causal Regulations in Non-Small Cell Lung Cancer.Front Oncol. 2021 Apr 27;11:655757. doi: 10.3389/fonc.2021.655757. eCollection 2021. Front Oncol. 2021. PMID: 33987090 Free PMC article.
-
A Phenotypic Approach to the Discovery of Potent G-Quadruplex Targeted Drugs.Molecules. 2024 Aug 1;29(15):3653. doi: 10.3390/molecules29153653. Molecules. 2024. PMID: 39125057 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

